Abstract
Impaired clearance of neurotoxic debris in the brain exacerbates neurologic disease and presents a promising therapeutic target. Pharmacologic therapies can enhance meningeal lymphatic clearance in preclinical models but may be limited by systemic toxicities or invasive administration. Here we report a low-intensity, focused ultrasound protocol that noninvasively clears pathogenic substances from the cerebrospinal fluid and brain interstitium in mice. Using two models of hemorrhagic stroke, we demonstrate that this protocol clears the cerebrospinal fluid and interstitium of blood cells, which accumulate in the deep cervical lymph nodes via meningeal lymphatics. The protocol directly modulates molecular processes, including mechanosensitive channels, to shift microglial phenotypes and astrocytic aquaporin localization to reduce neuroinflammation and neurocytotoxicity. In the intracerebral hemorrhage model, it improves behavioral outcomes and increases survival with greater efficacy than a pharmacologic benchmark. The protocol satisfies Food and Drug Administration safety guidelines, supporting clinical translatability. If demonstrated effective clinically, it may provide therapeutic benefit not only in hemorrhagic stroke but also in other neurologic disorders that involve impaired debris clearance.
Similar content being viewed by others
Main
Impairment of cerebrospinal fluid (CSF) circulation has been associated with pathogenesis in various neurologic disorders1,2,3. In particular, impaired clearance of dispersed debris in CSF and interstitial fluid promotes neuroinflammation and neurocytotoxicity, contributing to pathology and symptom severity in ischemic stroke, hemorrhagic stroke, traumatic brain injury, neurodegenerative diseases and other neurologic conditions1,2,3. Pharmacologic upregulation of meningeal lymphatics to enhance clearance of neurotoxic debris has demonstrated therapeutic potential in preclinical models4,5,6. However, despite promising preclinical data, currently there are no approved pharmacologic therapies that directly enhance intracranial fluid clearance. Concerns about experimental drug candidates relate to off-target effects, systemic toxicities, prolonged latency of therapeutic effects and limited translational feasibility of invasive administration protocols7,8,9. Meanwhile, surgical approaches, such as direct CSF drainage in subarachnoid hemorrhage (SAH)10 and hematoma evacuation in intracerebral hemorrhage (ICH)11, provide clinical benefit but are reserved for the most severely affected individuals, are invasive and do not directly correct the underlying molecular and cellular pathophysiology.
Focused ultrasound (FUS) has been shown to either enhance or suppress neuroinflammatory profiles in the brain, with outcomes largely dependent on sonication parameters and further influenced by coadministration of exogenous agents such as microbubbles12. Low-intensity FUS paired with microbubbles may disrupt the blood–brain barrier, which clears Alzheimer’s-associated plaques in preclinical models13 but also has sterile neuroinflammatory effects14 and raises safety concerns, given the risk of microbubble inertial cavitation as well as practical concerns in coordinating sonication while the microbubbles rapidly clear with a half-life of minutes12. Recently, a FUS protocol without microbubbles recapitulated similar behavioral effects in the same Alzheimer’s model, but clearance of plaque and neurotoxic substances was not shown, and the required ultrasound frequency presents clinical challenges for transcranial transmission15.
Low-intensity FUS is known to activate mechanosensitive ion channels, such as Piezo1, both in vitro and in animals in vivo16,17. Additionally, pharmacologic agonists of mechanosensitive channels in animal studies have shown promise for neurologic therapeutics development as they can dampen neuroinflammatory states18,19,20 and enhance glymphatic mediators, including meningeal lymphatic vessel sprouting21,22. However, in many cases these agents must be administered invasively, either intraventricularly or intracisternally, or over multiple weeks21, making for practical translational challenges. They may also carry risks of systemic toxicity23,24. None are currently approved for human administration. These findings suggest that if ultrasound protocols could be tailored to activate mechanosensitive ion channels, they could promote similar beneficial effects on neuroinflammation and glymphatic circulation while avoiding invasive administration and systemic toxicity risks.
We and others previously demonstrated that low-intensity FUS alone, without coadministration of exogenous agents, enhances CSF circulation25,26,27. Ultrasound improved the brain distribution of intrathecally administered drugs and facilitated the circulation of cisternally and intraparenchymally administered tracers25,26,27. Although these studies did not establish therapeutic efficacy in a neurological disease model, they indicate the potential of low-intensity FUS to clear pathogenic substances from the central nervous system and to achieve corresponding therapeutic effects.
In this study, we developed and rigorously validated a low-intensity transcranial FUS protocol aimed specifically at enhancing clearance of neurotoxic debris. We assessed the protocol’s safety and efficacy in two mouse models of hemorrhagic brain injury: SAH and ICH. We examined clearance of micron-scale hemorrhagic debris from both the CSF and interstitial compartments, changes in neuroinflammation and neurocytotoxicity and the underlying cellular and molecular mechanisms of the clearance effect. Our results show that ultrasound treatment enhances neurotoxic debris clearance, reduces neuroinflammation and neurocytotoxicity, improves functional behavioral outcomes and increases survival, with greater safety and efficacy than a pharmacologic benchmark. At the molecular level, we document dependence on mechanosensitive ion channel activation, with an ultrasound-induced shift in microglia to a homeostatic phenotype and with increased polarization of aquaporins in astrocytic vascular endfeet. If validated in clinical trials, this approach would offer a clinically feasible, nonpharmacologic, noninvasive therapeutic strategy for stroke and potentially other neurologic disorders that involve impaired clearance of neurotoxic debris.
Results
Ultrasonic debris clearance enhances intracranial hemorrhage clearance
To assess the utility of low-intensity transcranial FUS to clear neurotoxic debris from the CSF and interstitial brain compartments in stroke models, we first adapted to mice our prior protocol25 that used continuous 650-kHz ultrasound in rats, given the multitude of disease models developed for mice. Although our earlier protocol was effective, its ultrasound center frequency of 650 kHz presents challenges for translation to larger animals and humans given both the more prominent absorption by the skull (and therefore risk of unsafe heating) as well as the aberrating effect of the skull and human hair compared to lower frequencies like 200–250 kHz (refs. 28,29). We first confirmed that our prior finding of increased CSF–interstitial fluid mixing with ultrasound showed no appreciable reduction when the center frequency was decreased to 250 kHz (Extended Data Fig. 1a,b). Next, because at 250 kHz the ultrasound focus using this f = 1.0 focused transducer would affect much of the mouse brain at once (Fig. 1a,b), to prevent heat accumulation during application, we used a pulsed protocol with a duty cycle (25%) that we estimated would not yield substantial parenchymal heating based on the bioheat equation30 and simulation and experimental verification (Extended Data Fig. 2), while keeping consistent the duty cycle of ultrasound application at points of the brain that we used in our prior protocol25. Finally, we completed a parameter variation of the applied ultrasound pressure (Extended Data Fig. 1c) to yield a target ultrasound protocol for use in mice. These results showing that ultrasound increases brain parenchymal entry of CSF-dispersed tracers confirm our prior25 findings that low-intensity FUS increases CSF and interstitial fluid exchange.
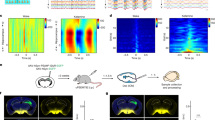
a, Schematic of the FUS protocol (250 kHz, 0.45-MPa peak negative pressure (PNP), 25% duty cycle, 50-ms pulse width) efflux experiment. b, Simulated ultrasound pressure beam profiles and effect of the skull on the intracranial profile. c, Timeline of SAH and ICH experiments. d, UDC clears CSF-dispersed red blood cells (RBCs) compared to sham in an SAH model (P = 0.0022). There is a greater accumulation of red blood cells in the deep cervical lymph nodes of SAH mice with UDC than in those of SAH sham mice (P = 0.0087). e, UDC clears interstitial red blood cells compared to sham in an ICH model (P = 0.0022). There is a greater accumulation of red blood cells in the deep cervical lymph nodes of ICH mice with UDC than in those with sham (P = 0.0411). No red blood cells were observed in separate cohorts of healthy control brain or lymph node tissue. Data presented as box plots that show minima, maxima, interquartile range (box bounds) and median (black line). Data were analyzed by two-tailed Mann–Whitney U-test; *P ≤ 0.05; **P ≤ 0.01; n = 6 for each group. Scale bars, 100 μm.
We next applied this ultrasonic debris clearance (UDC) protocol to two models of hemorrhagic brain injury in which impaired clearance of hemorrhagic products is thought to be pathogenic: SAH and ICH (also known as intraparenchymal hemorrhage)31,32. Additionally, the micron-scale hemorrhagic debris in these models provide an ambitious target for clearance, which, if successful, would suggest the potential to clear a wide variety of neurotoxic debris. In a mouse SAH model (Fig. 1c), UDC treatment reduced the number of red blood cells detected in the CSF compared to sham treatment (mean ± s.d.; SAH + sham: 338.33 ± 65.55 versus SAH + UDC: 140 ± 44.27, P ≤ 0.01; no extravascular red blood cells visible in healthy controls; Fig. 1d). This reduction was associated with a greater accumulation of red blood cells in the deep cervical lymph nodes, indicating efficient clearance through the meningeal lymphatic system (mean ± s.d.; SAH + sham: 12 ± 4.47 versus SAH + UDC: 24.83 ± 6.85, P ≤ 0.01; no extravascular red blood cells visible in healthy controls; Fig. 1d). Similarly, in the ICH model, UDC treatment reduced the number of interstitial red blood cells compared to sham (mean ± s.d.; ICH + sham: 26.17 ± 7.47 versus ICH + UDC: 11.67 ± 5.32, P ≤ 0.01; no extravascular red blood cells visible in healthy controls; Fig. 1e). This clearance was again accompanied by increased red blood cell accumulation in the deep cervical lymph nodes (mean ± s.d.; ICH + sham: 7.67 ± 4.37 versus ICH + UDC: 14.5 ± 4.32, P ≤ 0.05; no extravascular red blood cells visible in healthy controls; Fig. 1e). These findings demonstrate that UDC facilitates the clearance of micron-scale hemorrhagic debris from both the CSF and interstitial compartments to the deep cervical lymph nodes, likely through the meningeal lymphatics, arachnoid cuff exit points33 or arterial–venous leptomeningeal overlaps34 that are identified as main egress paths for CSF- and interstitial fluid-dispersed solutes.
UDC reduces posthemorrhagic neuroinflammation and neurocytotoxicity
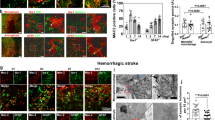
Neuroinflammation and neurocytotoxicity are critical factors that contribute to poor outcomes after hemorrhagic brain injury35. To evaluate the impact of UDC on these processes, we performed immunohistochemical analyses of brain tissue from UDC-treated SAH and ICH models (Fig. 2a,e). UDC treatment reduced the mean fluorescence intensity of IBA1+ cells (a commonly used marker of microglia, including both resting and activated states), suggesting an attenuation of microglial responses in both models (mean ± s.d.; healthy control: 18.5 ± 3.73; SAH + sham: 35 ± 8.27 versus SAH + UDC: 24 ± 7.62, ICH + sham: 44.7 ± 8.09 versus ICH + UDC: 30.5 ± 7.64, P ≤ 0.05; Fig. 2b,f). Additionally, astrocytic activation assessed by glial fibrillary acidic protein (GFAP) staining (an intermediate filament protein expressed by astrocytes that is strongly upregulated in reactive/activated astrocytes) was also reduced in UDC-treated animals in both models (mean fluorescence intensity ± s.d.; healthy control: 21.7 ± 3.85; SAH + sham: 43.33 ± 11.52 versus SAH + UDC: 24 ± 10.77, ICH + sham: 46 ± 8.39 versus ICH + UDC: 33.17 ± 9.06, P ≤ 0.05; Fig. 2c,g). These results indicate that UDC effectively attenuates neuroinflammatory responses after hemorrhagic brain injury. Furthermore, histological assessment of neuronal degeneration using Fluoro-Jade C staining (a fluorescent dye that selectively labels degenerating neurons regardless of the mechanism of cell death) revealed a reduction in the number of degenerating neurons in UDC-treated animals compared to sham (mean ± s.d.; healthy control: 2.0 ± 1.63; SAH + sham: 38.5 ± 9.54 versus SAH + UDC: 22.5 ± 7.40, ICH + sham: 35.5 ± 12.24 versus ICH + UDC: 17 ± 5.14, P ≤ 0.01; Fig. 2d,h). This reduction in neurotoxicity indicates the protective effects of noninvasive early blood clearance after hemorrhage in reducing neuronal damage.
a, Schematic of the SAH model with the location of interest (LOI) sampled for histological analyses. b,c, Immunohistological assessment of SAH brains for microglial activation (IBA1, purple; b) and gliosis/astrocytic activation (GFAP, green; c) revealed a decrease in both neuroinflammatory markers with UDC compared to sham (IBA1: P = 0.0411, GFAP: P = 0.0173); MFI, mean fluorescence intensity. d, Histological assessment of SAH brains for neuronal degeneration (Fluoro-Jade C (FJ-C; green) revealed a decrease with UDC compared to sham (P = 0.0087). e, Schematic of the ICH model with the LOI sampled for histological analyses. f,g, Immunohistological assessment of ICH brains for microglial activation (f) and gliosis/astrocytic activation (g) revealed a decrease in both neuroinflammatory markers with UDC compared to sham (IBA1: P = 0.0260, GFAP: P = 0.0455). h, Histological assessment of ICH brains for neuronal degeneration revealed a decrease with UDC compared to sham (P = 0.0043). Data presented as box plots that show minima, maxima, interquartile range (box bounds) and median (black line). Healthy control data are presented for reference and are repeated from b–d to f–h. Data were analyzed by two-tailed Mann–Whitney U-test; *P ≤ 0.05, **P ≤ 0.01; n = 6 for each group; AU, arbitrary units. Scale bars, 100 μm.
Improved behavioral outcomes and survival with UDC treatment
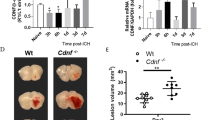
Next, we assessed functional recovery and survival as indicators of UDC efficacy. Our pilot studies indicated that the SAH model did not produce a sustained behavioral deficit. Therefore, we conducted behavioral, morbidity and mortality outcomes analyses using the ICH model (Fig. 3a). UDC treatment improved both the assayed behavioral and functional outcomes (corner turn test and grip strength) as early as 6 days after hemorrhage, with sustained improvements observed through 14 days (mean ± s.d.; from post-ICH to day 14: corner turn test in percent left limb use: healthy control baseline: 50.7 ± 5.33; ICH + sham: 26.89 ± 6.83 versus ICH + UDC: 39.13 ± 7.09; grip strength in force per g: healthy control baseline: 89.8 ± 7.91; ICH + sham: 61.67 ± 10.27 versus ICH + UDC: 77 ± 11.51, P ≤ 0.01 compared to sham; Fig. 3b).
a, Timeline of ICH outcomes experiment. b, Behavioral/functional assessments (corner turn test and grip strength) revealed an improvement in functional outcomes as early as 6 days after hemorrhage with UDC compared to sham (corner turn test, day 6: P = 0.0046, day 10: P = 0.0247, day 14: P = 0.0033; grip strength test, day 10: P = 0.0277, day 14: P = 0.0045). c, Decreased morbidity outcomes (body weight and brain water content, edema) with UDC compared to sham (body weight, day 14: P = 0.0035; brain water content P = 0.0108). d, Increased survival with UDC intervention compared to sham; χ2(2) = 4.08, P = 0.0432. Time series data points are presented as mean ± s.d., and two group comparisons presented as box plots show minima, maxima, interquartile range (box bounds) and median (black line); healthy control baseline levels are presented for reference (dashed lines, gray). Data in b and c were analyzed by Mann–Whitney U-tests with multiple comparisons controlled using the false discovery rate (FDR) method (two-stage step-up, Benjamini, Krieger and Yekutieli). Data in c were analyzed by Kruskal–Wallis H-test followed by a Dunn’s multiple comparisons test (left) or two-tailed Mann–Whitney U-test (right). Data in d were analyzed by log-rank (Mantel–Cox) test; *P ≤ 0.05, **P ≤ 0.01; n = 18 for each group; n = 6 for brain edema endpoint.
Morbidity outcomes, including body weight and brain water content (indicative of edema), were also assessed. UDC-treated mice exhibited lower brain edema and recovered their body weight faster than sham-treated controls (mean ± s.d.; brain edema, as percent brain water content: healthy control baseline: 72.6 ± 3.40; ICH + sham: 81.17 ± 2.40 versus ICH + UDC: 76.17 ± 3.37; body weight in grams at 14 days: healthy control baseline: 32.4 ± 3.10; ICH + sham: 26.33 ± 3.91 versus ICH + UDC: 31.07 ± 2.54, P ≤ 0.05; Fig. 3c). Notably, UDC treatment increased survival rates compared to sham (83.3% versus 50% at 14 days, P < 0.05; Fig. 3d). These disparate survival rates between the sham and UDC groups support the positive behavioral effects (Fig. 3a,b), which were observed despite a survivorship bias in these data that otherwise serves to lessen the observable behavioral effect sizes. These findings demonstrate that UDC not only enhances functional recovery but also reduces morbidity and mortality after hemorrhagic brain injury.
To provide a benchmark comparison, we tested a pharmacologic agonist of mechanosensitive channels (Yoda-1) using prior protocols that induced CSF clearance22. In the ICH model, these protocols proved detrimental, with decreased survival of treated mice compared to sham (Extended Data Fig. 3a,b). Mice that survived to the endpoint showed no decrease of hematoma volume, indicating reduced clearance efficacy compared to UDC (Extended Data Fig. 3c). Thus, UDC is safer and more efficacious than a benchmark pharmacologic therapy in this setting.
UDC induces homeostatic neuroimmunologic changes and cellular glymphatic processes
To investigate the potential cellular and molecular effects induced by UDC, we applied a spatial transcriptomic analysis to brain tissue from both healthy control and ICH model mice treated with sham or UDC (Fig. 4a). Consistent with prior literature and our assessment of activated microglia and astrocyte states (Fig. 2), the ICH model induced prominent upregulation of neuroinflammation and microglial activation (Fig. 4b). In the ICH model, UDC reduced both inflammation and microglial scores compared to sham, particularly in the perihematomal region, with a similar magnitude of transcriptional change of both disease-associated and homeostatic microglial states (Fig. 4b). Investigating specific factors of interest, we observed substantial transcriptional upregulation of Aqp4, Gfap, Cd68, Aif1 (IBA1), Lgals3, P2ry6 and P2ry14 in the ICH model, particularly in the perihematomal region. With UDC treatment of the ICH model, the neuroinflammatory genes Cd68 and Lgals3 showed relative downregulation in the perihematomal region (Fig. 4c), consistent with our prior analysis of reduced microglial activation with UDC (Fig. 2). By contrast, P2yr12, a homeostatic microglial marker, showed a trend toward increased expression with UDC treatment of ICH. For Aqp4, although transcript levels were not altered by UDC in the ICH model, we note that AQP4 is strongly expressed in astrocytic endfeet at the gliovascular interface, and changes in its protein localization or polarization may occur independently of transcriptional changes. At the transcriptional level, no definite change of Aqp4 or Gfap was noted with UDC in the ICH model, in potential contrast to the observed changes in expression of GFAP protein (Fig. 2c,g). No definite changes of these select markers were noted after UDC in healthy control animals. The largest activation in perihematomal gene expression was observed between ICH and control (1,777 genes upregulated and 19 genes downregulated in ICH). UDC restored ICH perihematomal gene expression toward its baseline, with substantial gene downregulation in ICH + UDC versus ICH (486 genes downregulated in ICH + UDC) and only modest gene expression differences between ICH + UDC and healthy control animals (466 genes upregulated and 14 genes downregulated in ICH + UDC). Additionally, UDC induced only limited gene expression changes in control animals (324 upregulated and 21 downregulated in control + UDC versus control), supporting the interpretation that its effects are specific to injury-related pathways (Fig. 4d). Together, these data suggest that UDC corrects ICH-associated gene networks, without substantial or concerning off-target gene activation in control conditions.
a, Brain region-level annotations of Leiden-derived spatial clusters of stereotranscriptomic data at a bin size of 100. Bins are colored by annotation; 4th ven., fourth ventricle; Cblm, cerebellum; ChP, choroid plexus; DG, dentate gyrus; DL-Ctx, deep-layer cortex; Hyp, hypothalamus; NAc–Str, nucleus accumbens-striatum; OB, olfactory bulb; RN–BS–Pons, reticular nucleus/brain stem/pons; Str, striatum; Thal, thalamus; UL-Ctx, upper-layer cortex; WM, white matter. The striatum and thalamus regions together constitute the perihematomal regions of the ICH groups. b, Gene group scoring across different brain regions and conditions (left: heat map; right: spatial distribution; inflammation score (GO:0006954) and microglia score (GO:1903978)); ‘disease-associated’ and ‘homeostatic’ scores are from manually curated lists (see Methods); Ctl, control. c, Key markers of glymphatics (Aqp4), glia (Gfap), microglia (Aif1, P2yr12) and inflammation (Cd68, Lgals3). d, Volcano plots of all genes across four different comparisons for data derived from the striatum and thalamus, which for ICH animals constitute the perihematoma region. The numbers of differentially expressed genes are shown in parentheses, determined as | log2 (fold change) | >1; q < 0.05.
To determine whether these transcriptional changes were reflected at the protein level, we studied an immunohistochemistry panel in the ICH model with and without UDC treatment (Fig. 5a). First, given our findings that UDC treatment of hemorrhagic brain injury reduced GFAP protein (Fig. 2c,g), but not Gfap mRNA (Fig. 4c), expression, we assessed the expression of the independent astrocytic marker S100β via immunohistochemistry, which showed no difference after UDC. This indicates that UDC does not reduce astrocyte numbers and that the decrease in GFAP protein expression after UDC may reflect either a biological discrepancy between Gfap mRNA and GFAP protein or a per astrocyte decrease of activation state (Fig. 5b)36,37. Next, following the spatial transcriptomics data suggesting changes in microglial state (Fig. 4), we assessed markers of different microglial states identified in the literature38,39, which include disease-associated microglia and homeostatic microglia. Microglial ramification branch complexity is a marker of microglial state, with high branch complexity correlating to the homeostatic state40,41. UDC increased microglial branch complexity (mean branch levels ± s.d.; ICH + sham: 3.68 ± 0.28 versus ICH + UDC: 5.23 ± 0.77, P ≤ 0.05; Fig. 5c). In agreement with this result and the spatial transcriptomics analysis, CD68 and GAL-3, which are markers of microglial activation42,43, were both decreased with UDC (mean number of cells ± s.d.; CD68: healthy control: 0.0 ± 0; ICH + sham: 401.5 ± 200.17 versus ICH + UDC: 45.63 ± 29.49, GAL-3: healthy control: 0.0 ± 0; ICH + sham: 727.5 ± 66.75 versus ICH + UDC: 257 ± 131.97, P ≤ 0.05; Fig. 5d). Likewise, P2RY12, a cell surface marker of homeostatic microglia necessary for the clearing of cellular debris44, increased with UDC (mean number of cells ± s.d.; healthy control: 25.5 ± 0.32; ICH + sham: 12.05 ± 3.84 versus ICH + UDC: 21.18 ± 0.95, P ≤ 0.05; Fig. 5e). Additionally, we observed a notable increase in the colocalization of red blood cell markers (TER-119+) with microglia (IBA1+) in the perihematoma region, suggesting elevated microglial phagocytosis of erythrocytes with UDC treatment of ICH (mean number of phagocytosed cells ± s.d.; ICH: 1.5 ± 0.58 versus ICH + UDC: 9.75 ± 2.5, P ≤ 0.05; Fig. 5f and Supplementary Video 1). Finally, in contrast to the overall transcriptomic upregulation of Aqp4 in ICH (Fig. 4c), we observed a nonstatistically significant trend toward decreased astrocytic endfeet perivascular polarization of AQP4 after ICH (Fig. 5g). AQP4 endfeet perivascular polarization, a key requirement of glymphatic upregulation, was restored in ICH models with UDC treatment (mean fluorescence intensity ± s.d.; healthy control: 37.15 ± 3.04; ICH + sham: 21.03 ± 7.53 versus ICH + UDC: 50.8 ± 11.80, P ≤ 0.05). Considering the totality of transcriptomic and immunohistochemistry data, we conclude that UDC decreases neuroinflammation via shifting microglia to the homeostatic phenotype and upregulates glymphatic function by increasing perivascular polarization of AQP4 in astrocytic endfeet.
a, ICH model with the LOI sampled for histological analyses. b, Immunohistological assessment of ICH brains for constitutive astrocytic counts (S100β+) revealed no significant difference with ICH + UDC compared to ICH + sham (P = 0.1455); FOV, field of view. c, Immunohistological assessment with three-dimensional (3D) reconstruction of microglia revealed increased branching, a hallmark of the homeostatic microglial state, with ICH + UDC compared to ICH + sham (P = 0.0286). Scale bars, 5 μm. d, Immunohistological assessment of microglial inflammatory states (CD68+, red; GAL-3+, yellow) revealed decreased proinflammatory disease-associated microglial markers with ICH + UDC compared to ICH + sham (CD68: P = 0.0286; GAL-3: P = 0.0286). CD68+ scale bars, 50 μm; GAL-3+ scale bars, 100 μm. e, Immunohistological assessment of purinergic receptors (P2RY12+, yellow) that reflect a homeostatic microglial state revealed an increase with ICH + UDC with a trend toward the healthy controls compared to ICH + sham (P = 0.0286). Scale bars, 20 μm. f, Immunohistological assessment with 3D reconstruction of microglia (IBA1+, green) revealed increased phagocytosis of red blood cells (TER-199+, red) with ICH + UDC compared to ICH + sham (P = 0.0286). Scale bars, 5 μm. g, Immunohistological assessment of astrocytic endfeet water channel localization (AQP4+, green) revealed increased polarization to the vasculature with ICH + UDC compared to ICH + sham (P = 0.0286). Data presented as box plots that show minima, maxima, interquartile range (box bounds) and median (black line). Data were analyzed by two-tailed Mann–Whitney U-test; *P ≤ 0.05; n = 4 for each group.
Ultrasound-mediated debris clearance acts through molecular mechanotransduction pathways
To further investigate the underlying mechanisms of ultrasound-mediated clearance, we considered potential thermal effects (Extended Data Fig. 2). At the intensity, duty cycle and ultrasound frequency used here, computational simulations (via k-Wave45) indicated no substantive parenchymal heating, with heat accumulation being minimal and preferentially in the skull, not the parenchyma. We confirmed this experimentally by measuring brain temperature before and after protocol application with a thermocouple placed intracranially, immediately under the skull. We noted a cooling trend over the experiment, presumably due to thermal conduction into the room temperature gel and water used for ultrasound coupling in contact with the dorsal scalp (peak intracranial temperature change: –0.5 ± 0.2 °C; Extended Data Fig. 2).
Given this result, we considered possible mechanical mechanisms such as direct induction of convection or oscillation of the intracranial fluid medium that promotes exchange of the CSF and interstitial fluid (as in Extended Data Fig. 1)25, with or without an accompanying mechanical change of the parenchyma that promotes downstream CSF circulation46. However, our prior results demonstrated that the response to UDC extends for over 1 h after ultrasound exposure25, implying a more prolonged mechanism. Because the ultrasound intensity of our protocol would be sufficient to activate mechanosensitive channels16,17, which can induce meningeal lymphatic vessel sprouting and increased CSF egress21,22, we investigated the effects of UDC on intracranial mechanosensitive channels. Using the ICH model, we cisternally administered a broad-spectrum mechanosensitive ion channel blocker (GsMTx4, a selective blocker of Piezo and TRP family channels; Fig. 6a) and observed elimination of the blood cell clearance efficacy of UDC, as evidenced by increased hematoma volumes in the UDC + GsMTx4 treatment group versus UDC treatment alone (mean hematoma area percent ± s.d.; ICH + UDC: 11.63 ± 4.59 versus ICH + UDC + GsMTx4: 23.63 ± 5.50) and no difference between the sham + GsMTx4 and UDC + GsMTx4 groups (Fig. 6b). The elimination of detectable clearance with GsMTx4 blockade (Hedge’s g, corrected for N ≤ 50 = 2.96 for sham versus UDC alone compared to 0.46 for sham + GsMTx4 versus UDC + GsMTx4) indicates that mechanosensitive ion channels mostly mediate the clearance effects of UDC. We observed no change in meningeal lymphatic vessel coverage or caliber in the ICH model or in response to UDC treatment (Fig. 6c), potentially reflecting the short, days-long timeline of both this disease model and the intervention. By contrast, we observed that UDC, when applied only once to healthy control brains, acutely increased P2RY12+ microglia, a marker of the homeostatic state44 (mean fluorescence intensity ± s.d.; healthy controls: 25.5 ± 0.37 versus healthy controls + UDC (one time): 27.98 ± 0.97, P ≤ 0.05; Fig. 6d), supporting the role of mechanosensitive channels in UDC efficacy. Notably, AQP4 expression did not change in healthy control brains after only one application of UDC (Fig. 6e), suggesting that UDC acts indirectly to drive the observed glymphatic enhancement in ICH (Fig. 5g). Instead, AQP4 polarization with UDC treatment of ICH may be secondarily induced through more direct UDC-driven reductions in neuroinflammation, with secondary disinhibition of AQP4 polarization, suggesting a distinction between the direct immunomodulatory effects of UDC and its relatively indirect glymphatic effects of AQP4 polarization.
a, Timeline of ICH experiment with UDC and a mechanosensitive ion channel blocker (GsMTx4). b, Administration of GsMTx4 reduced the blood product clearance of UDC, as assessed by hematoma volumes across treatment groups. c, Immunohistological analysis of meningeal lymphangiogenesis (LYVE-1+) revealed no significant difference between control, ICH + sham and ICH + UDC during this timeline of injury and treatment; SSS, superior sagittal sinus; TS, transverse sinus; NS, not significant. d, In healthy, uninjured mice, immunohistological analysis of purinergic receptors (P2RY12+, yellow) that reflect a homeostatic microglial state revealed an acute increase with only one application of UDC compared to control (P = 0.0286). e, In healthy, uninjured mice, immunohistological analysis of AQP4 (green) at astrocytic endfeet revealed no difference in polarization with one application of UDC compared to control (P = 0.6286). Data presented as box plots that show minima, maxima, interquartile range (box bounds) and median (black line); healthy control data are presented for reference. Data were analyzed by Kruskal–Wallis test with a post hoc Dunn’s multiple comparisons test; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; n = 8 mice for each group in Fig. 6b and n = 4 mice for each group in c–e.
Discussion
We have demonstrated that our UDC protocol facilitates the clearance of cellular debris from the mouse brain, reduces neuroinflammation and neurocytotoxicity and improves functional outcomes and survival after hemorrhagic brain injury (Figs. 1–3). UDC was safer and more efficacious than a pharmacologic benchmark for this indication (Extended Data Fig. 3). Our mechanistic studies suggest (Extended Data Fig. 4) that the clearance effects of UDC depend primarily on activation of mechanosensitive channels (Fig. 6a,b) and increased intracranial fluid compartmental intermixing (Extended Data Fig. 1)25. Activation of mechanosensitive channels shifts the microglial phenotype to the homeostatic state (Figs. 5c–e and 6d), promoting debris clearance in both the CSF and interstitial compartments (Figs. 1 and 5f and Supplementary Video 1). This shifting neuroinflammatory milieu promotes increased astrocytic endfeet perivascular polarization (Fig. 5g) by disinhibiting polarization in the setting of increased aquaporin expression after hemorrhage (Fig. 4). Our data suggest a model (Extended Data Fig. 4) in which decreased neuroinflammation, increased fluid compartment intermixing and increased glymphatic mediators, like aquaporin astrocytic endfeet perivascular polarization, combine to clear debris from the central nervous system to the lymphatic system of the body (Fig. 1), which in turn lowers morbidity and mortality in hemorrhagic stroke models (Figs. 2 and 3).
It should be emphasized that this model is still hypothetical, and several aspects require further investigation and validation in future studies. For instance, the mechanosensitive channel antagonist GsMTx4, which blocks UDC efficacy (Fig. 6a,b), limits mechanotransduction at the level of the lipid bilayer and therefore antagonizes the broad general class of mechanosensitive channels, including most, if not all, of the Piezo and TRP families47,48, making it unclear which specific channels (Piezo1, Piezo2, TRPA1, TRPC6 and so on) most contribute to UDC efficacy. It is also worth noting that we did not observe changes in lymphatic vessel coverage or morphology with UDC (Fig. 6c), although mechanosensitive channel activation can stimulate lymphatic flow without lymphatic morphological change21. Additionally, as mechanical stimulation of the face and neck can increase lymphatic drainage of the central nervous system49 and UDC is a form of mechanical stimulation, this mechanism may partly contribute to UDC efficacy.
We designed our UDC protocol to be compatible with clinical translation. The 250-kHz ultrasound frequency we used is in the range of commercial clinical transcranial FUS systems on the market worldwide50. This ultrasound frequency has relatively low attenuation and aberration through the human scalp and skull28,29. Additionally, the target in situ peak negative pressure we used corresponds to a mechanical index (a metric indicating the risk of mechanical injury induced by ultrasound) of 0.9, which is well within Food and Drug Administration guidelines for safe application51 and similar to that of recent studies of ultrasound neuromodulation51,52. This suggests that clinical trials of UDC should receive a nonsignificant risk designation from local institutional review boards.
If debris clearance by UDC is validated in clinical trials, it would not incur the risks of invasive surgical approaches nor the systemic toxicities of pharmacologic treatments and would not require coadministration of exogenous agents like nanoparticles or microbubbles53, simplifying clinical application and increasing its safe operating regime. As its mechanism does not require a particular pattern of brain activity (that is, it is effective in anesthetized animals), it would be applicable in humans who are in a range of cognitive states, including awake, asleep, engaged in a task or obtunded, in the immediate postinjury recovery period. Furthermore, it would provide a method to drive CSF circulation and egress complementary to pharmacologic approaches and approaches that activate particular neural activity patterns that are correlated with this circulation54,55. Accordingly, it would offer an alternate method for understanding and intervening in glymphatic CSF circulation and egress in the variety of disorders and normal states in which it has been implicated, including sleep56, neurodegeneration57, traumatic brain injury58 and mental health conditions59.
Methods
Animals
All experiments were approved by the Institutional Animal Care and Use Committees of Stanford University. Experiments were conducted with adult (3- to 4-month old) and aged (18- to 20-month old) male C57BL/6NCrl mice with a body weight of 30–40 g (Charles River Laboratories). All mice were housed in a temperature- (22 °C) and humidity-controlled (33–39%) environment under a 12-h/12-h light/dark cycle and were provided with food and water ad libitum. Mice were randomly assigned to one of two treatment groups: (1) no treatment (sham) and (2) treatment with a FUS protocol, herein referred to as UDC.
UDC protocol
FUS (250-kHz center frequency, 0.45-MPa peak in situ negative pressure, 50-ms pulse width, 25% duty cycle for 10 min via a 250-kHz center frequency, 70- or 100-mm aperture, f = 1.0) or sham (ultrasound power off) was applied transcranially throughout the brain (Extended Data Fig. 2). Before application, the fur on the head was removed using chemical hair depilatory. The transducer was coupled with ultrasound gel to the dorsal surface of the head, and ultrasound was applied while the mice were anesthetized under ketamine/xylazine (90 mg per kg (body weight) and 10 mg per kg (body weight), respectively). Body temperature, cardiac and respiratory rates and O2 saturation were monitored throughout the experiment. Environmental heating was used to help maintain body temperature.
Hemorrhagic models
Two previously characterized models of hemorrhagic stroke, SAH60 and ICH61, were implemented in this study. Briefly, for the SAH model, 25 µl of autologous blood was withdrawn from the tail vasculature to heparinized capillary tubing and injected into the cisterna magna. For the ICH model, 25 µl of autologous blood was withdrawn from the tail vasculature and injected into the right striatum. Mice were anesthetized under ketamine/xylazine (90 mg per kg (body weight) and 10 mg per kg (body weight), respectively). Body temperature, cardiac and respiratory rates and O2 saturation were monitored throughout the experiment. Environmental heating was used to help maintain body temperature.
Histology and immunostaining
Mice were killed and fixed via transcardial perfusion with PBS and 4% paraformaldehyde at time points described in each experimental timeline (Figs. 1–6). Brains were fixed in 4% paraformaldehyde for 24 h, cryoprotected by serial incubation in 15% and 30% sucrose solutions for 24 h each and frozen at −80 °C in optimal cutting medium (OCT) compound. Brains were sectioned at a thickness of 30 μm using a cryostat (LEICA CM 1950). Every fifth section (150 μm apart) was collected for imaging. The specimen temperature was set at −21 °C. Tissue sections were stored in cryoprotectant solution before staining. For immunofluorescence, the free-floating tissue sections were blocked in 0.3% Phosphate Buffered Saline with Tween-20 with 10% normal goat serum for 1 h at room temperature and incubated with primary antibodies overnight at 4 °C. After washing with PBS three times for 5 min each, sections were incubated in secondary antibodies for 2 h at room temperature. The primary antibodies used in immunofluorescence included rabbit anti-LYVE-1 (1:500; Abcam, ab14917), rat anti-TER-119 (1:500; Invitrogen, 14-5921-82), recombinant rabbit anti-IBA1 (1:500; Abcam, ab178846), chicken anti-GFAP (1:500; Abcam, ab4674), recombinant rabbit anti-aquaporin 4 (1:500; Abcam, ab282586), rat anti-CD68 (1:400; Bio-Rad, MCA1957), rabbit anti-P2RY12 (1:200; Novus Biologicals, NBP2-33870) and rat anti-CD31 (1:200; BD Pharmingen, 550300). The following corresponding secondary antibodies were used: AlexaFluor 488-labeled goat anti-rabbit (1:500; Thermo Fisher Scientific, A32731), AlexaFluor 555-labeled goat anti-rat (1:500; Thermo Fisher Scientific, A48263), AlexaFluor 647-labeled goat anti-rabbit (1:500; Thermo Fisher Scientific, A32733) and AlexaFluor 488-labeled goat anti-chicken (1:500; Thermo Fisher Scientific, A32931). For neurodegeneration staining, tissue sections were incubated in 0.06% potassium permanganate for 20 min and in 0.0001% Fluoro-Jade C (Biosensis). Tissue sections were mounted on microscope glass slides (Fisher) and cover-slipped with ProLong Gold Antifade Mountant (Thermo Fisher Scientific). All images were collected with a fluorescence microscope (BZ-X800, Keyence) and processed with BZ-X Advanced Analysis Software (Keyence) or ImageJ Software (version 1.53, National Institutes of Health). For hematoma volume, the mean area of blood was calculated in coronal segments using ImageJ and reported as percent hematoma area over total hemispheric area. For immunofluorescence quantification involving fluorescence intensity, signals above manually thresholded background were used for region of interest segmentation to calculate total mean fluorescence intensity using BZ-X Advanced Analysis software and/or Bitplane IMARIS software (version 9.8.0). To quantify microglial branching complexity from IBA1-immunoreactive cells in confocal z-stack images, we used Bitplane IMARIS software (version 9.8.0). The ‘Modeling’ and ‘Filaments’ modules were used to reconstruct IBA1+ microglia, focusing on cells with processes extending from the soma. IMARIS’s automated cell detection algorithm was applied to identify all microglia within each image. Detected structures were manually reviewed to ensure accuracy, and any misidentified cells were corrected to refine the detection parameters before quantitative analysis. For analyses of phagocytosis, 3D models of microglia and red blood cells were generated using IMARIS software based on IBA1 and TER-119 immunostaining, as previously described62. Internalization and colocalization events between microglia and red blood cells were subsequently measured. To quantify AQP4 polarization, we assessed the spatial relationship between AQP4 and blood vessels using dual immunostaining for AQP4 and CD31 (a vascular endothelial marker). Analysis was conducted at ×40 in the caudate–putamen region and the thalamus. Using Imaris software, we isolated the AQP4 and CD31 channels. A vascular mask was generated from the CD31 signal, and the shortest distance (0–0.2 µm) between AQP4+ voxels and CD31-labeled vessels was measured. The degree of AQP4 polarization was quantified based on the proportion of AQP4 signal within this 0- to 0.2-µm perivascular zone, reflecting localization at astrocytic endfeet. To quantify meningeal lymphatic percent coverage and vessel diameter, we performed immunostaining for LYVE-1 (a lymphatic endothelial marker). Analyses were conducted in the dorsal meninges overlying the superior sagittal sinus and transverse sinus. Confocal z stacks were acquired at ×20 magnification and processed using ImageJ (Fiji distribution). LYVE-1 and TER-119 channels were separated, and maximum intensity projections were generated for each region. To assess lymphatic coverage, a binary mask of the LYVE-1 signal was created using automatic thresholding. The percent coverage was calculated as the ratio of LYVE-1+ area to the total meningeal region of interest, which was manually outlined based on anatomical boundaries and TER-119 signal. For vessel diameter quantification, the LYVE-1 binary mask was skeletonized using the ‘Skeletonize’ function, and local diameters were measured using the ‘Analyze Skeleton’ and ‘Local Thickness’ plugins. Values were averaged across multiple fields per animal and sampled from the superior sagittal sinus and transverse sinus regions. All imaging and thresholding parameters were completed by personnel blinded to the experimental conditions.
Neurobehavioral function evaluation
Two behavioral tests were used to evaluate behavioral and functional outcomes as previously described: a corner turn test and a grip strength test63. Briefly, for the corner turn test, quantification of turning preference after approaching a 30° corner was used to assay sensorimotor deficits. The values were calculated as the percentage of left versus right limb use on a turn for a total duration of 3 min per session. For the grip strength test, motor function was assessed via the peak force (G) required for mice to release their grip from a grid bar as quantified by a digital grip strength meter (Maze Engineers). The average of three attempts was calculated per test session. All behavioral assessments were completed by personnel blinded to experimental conditions. All animals were subjected to stroke induction before randomization into treatment and control groups, ensuring that baseline injury severity was equally distributed.
Morbidity evaluation
Total body weight (g) and brain water content (edema) percentage were used as indicators of posthemorrhagic morbidity. Brain water content was assessed at day 14 after hemorrhage. The wet weight of each brain was recorded after euthanasia and extraction. The brains were then dehydrated at 110 °C for 72 h. Dry weights were recorded, and brain edema was evaluated as the difference in percent brain water content.
Reagents
For mechanosensitive ion channel inhibition, we mixed GsMTx4 (MedChem Express, HY-P1410) with artificial CSF and injected (5 μM in 5 μl) it into the cisterna magna of mice 0.5 h before FUS application. For mechanosensitive ion channel activation, we mixed Yoda-1 (Sigma-Aldrich) in DMSO at 710 μg ml−1 and diluted it in PBS at a 3:100 (vol:vol) ratio of DMSO to PBS. The final solution was administered at a dosage of 213 μg per kg (body weight) intraperitoneally per day, following prior studies22.
Tissue collection and processing for Stereo-seq
Animals were killed on day 6 (after ICH; see the experimental timeline in Fig. 1c), and brain tissues were extracted and snap-frozen in isopentane prechilled in liquid nitrogen in Tissue-Tek OCT (Sakura, 4583) and transferred to a –80 °C freezer for storage. Cryosections were cut sagittally at a thickness of 10 μm on a Leica CM1860 cryostat. Sections were matched to include equivalent regions of olfactory bulb, striatum, lateral and fourth ventricles, thalamus, midbrain and cerebellum. Stereo-seq experiments were performed as previously described64 and followed STOmics Stereo-seq Chip-on-a-Slide (1 cm × 1 cm) v1.3 kit instructions, with minor modifications. Cryosections were melted onto Stereo-seq T-chips that had been rinsed with deionized water and dried with dust off by placing the section on the chip using tweezers at –20 °C in a cryostat and removing the slide from the cryostat. Afterward, slides were baked for 5 min at 37 °C and submerged in prechilled methanol at –20 °C for 30 min. After drying, the slides were washed one time in 0.1× SSC and placed in a gasket to enable easy addition and removal of solution directly to the chip. In total, 150 μl of prewarmed 1× permeabilization solution was added to the gasket, and chips were incubated at 37 °C in a thermocycler for 12 min. The reagent was then removed, washed once with 0.1× SSC, 200 μl of 1× reverse transcriptase solution was added, and chips were incubated for 2 h at 45 °C. After removing the reverse transcriptase solution, chips were again washed once with 0.1× SSC, and then 180 μl of cDNA release solution was added and incubated for 10 min at 55 °C. The solution was then mixed thoroughly to remove sample from the chip and collected into a new tube. We added 23 μl of neutralization buffer to the tube, split the sample into three PCR tubes and incubated at 95 °C for 5 min. Afterward, we amplified the cDNA with 34 μl of PCR mix and ran the PCR using the following settings: 95 °C for 5 min, 13 cycles of 98 °C for 20 s, 58 °C for 20 s and 72 °C for 3 min, followed by 72 °C for 5 min. Afterward, we performed 1:0.8 bead cleanup on samples using AMPure beads following the manufacturer’s protocol.
Library construction and sequencing of Stereo-seq data
Libraries were constructed and barcoded using a STOmics barcode library preparation kit v1.0. We fragmented 300 ng of amplified cDNA with 10 μl of KMB in a total reaction volume of 45 μl and incubated in a thermocycler at 95 °C for 5 min and 40 °C for 3 min. The reaction was stopped by adding 5 μl of KME and incubated at 37 °C for 10 min. Fragmented DNA was cleaned up using AMPure beads with a ratio of 1:0.8 beads following the manufacturer’s protocol. DNA was eluted in 25 μl of 1× TE, combined with 25 μl of a unique barcode mix and 50 μl of 2× PCR master mix, and amplified using the following program: 95 °C for 5 min, 13 cycles of 98 °C for 20 s, 58 °C for 20 s and 72 °C for 30 s, followed by 72 °C for 5 min. The final product was cleaned up using a double-sided AMPure bead cleanup with a 1:0.55 bead ratio following the manufacturer’s protocol. Libraries were eluted in 20 μl of 1× TE. Prepared libraries were then sequenced using a DBSEQ-T7RS sequencer at the Stanford Genomics Core, targeting 1.5 billion 75-base pair (bp) paired-end reads per sample.
Processing of Stereo-seq raw data
FASTQ files corresponding to barcode indexes per sample along with the DNA nanoball (DNB) mask file were processed using the SAW v8.1.3 count pipeline65. The sequences from the FASTQ files were filtered to only include reads with one or fewer unmapped base pairs in the cellular indexing domain (CID). CIDs were then mapped to the corresponding DNB mask file, and valid CIDs were associated with reads and retained for subsequent steps. Next, FASTQ files were filtered to only include reads that contained >30 bp not including adapter, DNB or poly(A) sequences. Afterward, sequences with low-quality molecular identified sequences were removed (quality of ≤Q10 or unmapped base pairs of ≥1). A reference file for GRCm39vM30 was assembled, and reads were aligned using STAR66. We used default parameters for the SAW count pipeline, which uses STAR and performs alignment, annotation, molecular identified correction and deduplication and binning. The outputted gene expression file contained the spatial gene expression matrix at different bin sizes.
Analysis of Stereo-seq data
The resulting gene expression files for all four samples were loaded into a multisample data object (MSdata) at a bin size of 100 (50 μm × 50 μm) using Stereopy67 and integrated. Using Stereopy default parameters, the expression data were log normalized per bin. Highly variable genes were calculated, a principal component analysis was performed on highly variable genes, and Harmony68 was run to minimize batch effects in the embedding space. Afterward, nearest neighbors were calculated using the first 50 principal components, uniform manifold approximation and projection images were constructed, and Leiden was run at a resolution of 2. Leiden clustering was further refined by running Stereopy’s spatial neighbors algorithm and performing Leiden again at a resolution of 2. Clusters containing fewer than 300 bins across all samples were left unannotated, and remaining clusters were annotated based on spatial location, known marker genes and previously annotated Stereo-seq data69.
After annotation, the MSdata object was converted into annotated data (anndata) objects and analyzed using Scanpy70. Differentially expressed genes within the perihematomal region (striatum + thalamus) were found using the Scanpy rank_gene_groups function using method = ‘wilcoxon’, and differentially expressed genes were selected using | log2 (fold change) | of >1 and adjusted P value of <0.05 as thresholds. Gene scores were calculated using the Scanpy score_genes function based on gene lists for inflammation (GO:0006954) or microglial activity (GO:1903978) or disease-associated microglial and homeostatic microglial scores from manually curated lists of genes (disease-associated: ‘Ifng’, ‘Tnf’, ‘Il6’, ‘Il1b’, ‘Il12a’, ‘Il12b’ ‘Ccl2’, ‘Cybb’, ‘Nos2’, ‘H2-Aa’, ‘H2-Ab1’, ‘Itgam’, ‘Itgax’, ‘Cd36’, ‘Ptprc’, ‘Cd47’, ‘Fcgr1’, ‘Fcgr2b’, ‘Fcgr3’, ‘Fcgr4’ and ‘Fcer1g’; homeostatic: ‘Il4’, ‘Il13’, ‘Il10’, ‘Tgfb1’, ‘Igf1’, ‘Fgf2’, ‘Csf1’, ‘Ngf’, ‘Bdnf’, ‘Ntf3’, ‘Gdnf’, ‘Grn’, ‘Mrc1’, ‘Retnla’, ‘Chil3’ and ‘Arg1’). Spatial plots of annotations, gene scores and individual genes were generated using Squidpy71.
Thermal evaluation
A male C57BL/6 mouse (weight: 35 g) was imaged on a Quantum GX micro-CT to simulate the pressure field distributions during FUS treatment. The obtained micro-CT image was 474 × 504 × 921 with a cubic voxel size of 0.0499 mm. The images were resampled linearly to a cubic voxel size of 0.34 mm. The bone, soft tissue and water were isolated based on their Hounsfield units (1,200 HU for bone/soft tissue and 930 HU for soft tissue/water threshold). Density and sound speed were linearly interpolated in each region using hounsfield2density, a predefined function through the k-Wave MATLAB toolbox45. The region surrounding the animal was defined as water. The transducer was described as a bowl with a diameter and radius of curvature of 100 mm. At a center frequency of 250 kHz, the points per wavelength was 17.44 in water and Courant–Friedrichs–Lewy stability criterion of 0.1, leading to a time step of 22.8 ns. The simulation was run for 85 µs, allowing the initial wave to travel to the length of the simulation grid and back past the top of the skull (135 mm). All simulations were conducted using the k-Wave toolbox45. After the pressure field was complete, the time array was cropped to remove the expected travel time from the transducer surface to the top of the skull (60 to 85 µs). The field of view was reduced to double the focal region of the ultrasound (full-width at half-maximum) and double the height of the skull. The resulting simulation space was 15 × 15 × 20 mm for 25 µs. The phase and pressure amplitude were extracted using the built-in k-Wave function extractAmpPhase. The heat disposition was calculated for each voxel using the original density, speed of sound and attenuation coefficient determined previously. The starting temperature, thermal conductivity and specific heat were set to 37 °C, 0.51 W (m × K)−1 and 3,630 J (kg × K)−1, respectively. Tissue properties were obtained from the IT’IS database72. Thermal heating was determined for a single burst (50 ms on, 150 ms off) with a time step of 0.1 ms.
Statistical analysis
Sample size determination was guided by a power analysis performed using G*Power 3.1 software (Heinrich-Heine-Universität Düsseldorf). Parameters used for the analysis included an effect size (Cohen’s d) ranging from 0.8 to 2.0, an α error probability of 0.05, power (1 – β) of 0.8 and an allocation ratio (N2:N1) of 1. Effect sizes were estimated based on preliminary pilot data. All experiments were randomized. Data collection and analysis were conducted by investigators blinded to treatment groupings. Data normality was assessed using the Kolmogorov–Smirnov test where appropriate. Based on the outcomes and given respective sample sizes, nonparametric statistical methods were applied. For comparison of two groups, a two-tailed Mann–Whitney U-test (Wilcoxon rank-sum test) was used. For comparisons among multiple groups, a Kruskal–Wallis H-test followed by a Dunn’s multiple comparisons test was used. For repeated measures, multiple Mann–Whitney U-tests were corrected using the FDR method (Benjamini, Krieger and Yekutieli) with a desired FDR of 1.00% to assess statistical significance. Effect sizes were calculated using Hedges’ g to compare hematoma volume between groups, accounting for small sample sizes. The correction factor for Hedges’ g was applied to adjust for bias in small samples (N < 50), as previously described73. All statistical analyses were performed using GraphPad Prism 10.3.0 (GraphPad Software) and JMP Pro, V15.0 (SAS Institute). All data are presented as box plots (presenting all data points, the median, box limits of the interquartile range and whisker tails of the full data range or 1.5 times the interquartile range) or mean ± s.d. unless otherwise specified.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The spatial transcriptomics dataset generated and analyzed during the current study has been deposited in the Gene Expression Omnibus74 under accession number GSE296613. Any additional data not presented is available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Shah, V. A. et al. One-year outcome trajectories and factors associated with functional recovery among survivors of intracerebral and intraventricular hemorrhage with initial severe disability. JAMA Neurol. 79, 856–868 (2022).
Panni, P. et al. Hemorrhage volume drives early brain injury and outcome in poor-grade aneurysmal SAH. Am. J. Neuroradiol. 45, 393–399 (2024).
Perel, P. et al. Intracranial bleeding in patients with traumatic brain injury: a prognostic study. BMC Emerg. Med. 9, 15 (2009).
Chen, J. et al. Meningeal lymphatics clear erythrocytes that arise from subarachnoid hemorrhage. Nat. Commun. 11, 3159 (2020).
Tsai, H.-H. et al. Functional investigation of meningeal lymphatic system in experimental intracerebral hemorrhage. Stroke 53, 987–998 (2022).
Wang, X. et al. Dobutamine promotes the clearance of erythrocytes from the brain to cervical lymph nodes after subarachnoid hemorrhage in mice. Front. Pharmacol. 13, 1061457 (2023).
Dela Paz, N. G. & Frangos, J. A. Yoda1-induced phosphorylation of AKT and ERK1/2 does not require Piezo1 activation. Biochem. Biophys. Res. Commun. 497, 220–225 (2018).
Wijerathne, T. D., Ozkan, A. D. & Lacroix, J. J. Yoda1’s energetic footprint on Piezo1 channels and its modulation by voltage and temperature. Proc. Natl Acad. Sci. USA 119, e2202269119 (2022).
PubChem compound summary for CID 2746822, Yoda-1. National Center for Biotechnology Information https://pubchem.ncbi.nlm.nih.gov/compound/2746822 (2025).
Wolf, S. et al. Effectiveness of lumbar cerebrospinal fluid drain among patients with aneurysmal subarachnoid hemorrhage: a randomized clinical trial. JAMA Neurol. 80, 833–842 (2023).
Pradilla, G. et al. Trial of early minimally invasive removal of intracerebral hemorrhage. N. Engl. J. Med. 390, 1277–1289 (2024).
Leinenga, G., Langton, C., Nisbet, R. & Götz, J. Ultrasound treatment of neurological diseases—current and emerging applications. Nat. Rev. Neurol. 12, 161–174 (2016).
Leinenga, G. & Götz, J. Scanning ultrasound removes amyloid-β and restores memory in an Alzheimer’s disease mouse model. Sci. Transl. Med. 7, 278ra33 (2015).
Jung, O. et al. Neuroinflammation associated with ultrasound-mediated permeabilization of the blood–brain barrier. Trends Neurosci. 45, 459–470 (2022).
Leinenga, G. et al. Correction: scanning ultrasound-mediated memory and functional improvements do not require amyloid-β reduction. Mol. Psychiatry 29, 3968–3970 (2024).
Yoo, S., Mittelstein, D. R., Hurt, R. C., Lacroix, J. & Shapiro, M. G. Focused ultrasound excites cortical neurons via mechanosensitive calcium accumulation and ion channel amplification. Nat. Commun. 13, 493 (2022).
Zhu, J. et al. The mechanosensitive ion channel Piezo1 contributes to ultrasound neuromodulation. Proc. Natl Acad. Sci. USA 120, e2300291120 (2023).
Zhang, M., Malko, P. & Jiang, L.-H. Mechanosensitive Piezo1 channel regulation of microglial cell function and implications to neurodegenerative diseases and neuroinflammation. Neural Regen. Res. 18, 2385–2386 (2023).
Ikiz, E. D., Hascup, E. R., Bae, C. & Hascup, K. N. Microglial Piezo1 mechanosensitive channel as a therapeutic target in Alzheimer’s disease. Front. Cell Neurosci. 18, 1423410 (2024).
Zhu, T. et al. The mechanosensitive ion channel Piezo1 modulates the migration and immune response of microglia. iScience 26, 105993 (2023).
Choi, D. et al. Piezo1 regulates meningeal lymphatic vessel drainage and alleviates excessive CSF accumulation. Nat. Neurosci. 27, 913–926 (2024).
Matrongolo, M. J. et al. Piezo1 agonist restores meningeal lymphatic vessels, drainage, and brain–CSF perfusion in craniosynostosis and aged mice. J. Clin. Invest. 134, 559761 (2024).
Kim, Y.-J. & Hyun, J. Mechanosensitive ion channels in apoptosis and ferroptosis: focusing on the role of Piezo1. BMB Rep. 56, 145–152 (2023).
Romac, J. M.-J., Shahid, R. A., Swain, S. M., Vigna, S. R. & Liddle, R. A. Piezo1 is a mechanically activated ion channel and mediates pressure induced pancreatitis. Nat. Commun. 9, 1715 (2018).
Aryal, M. et al. Noninvasive ultrasonic induction of cerebrospinal fluid flow enhances intrathecal drug delivery. J. Control. Release 349, 434–442 (2022).
Yoo, S.-S. et al. Enhancement of cerebrospinal fluid tracer movement by the application of pulsed transcranial focused ultrasound. Sci. Rep. 12, 12940 (2022).
Wu, C.-H. et al. Very low-intensity ultrasound facilitates glymphatic influx and clearance via modulation of the TRPV4–AQP4 pathway. Adv. Sci. 11, 2401039 (2024).
Eames, M. D., Hananel, A., Snell, J. W., Kassell, N. F. & Aubry, J.-F. Trans-cranial focused ultrasound without hair shaving: feasibility study in an ex vivo cadaver model. J. Ther. Ultrasound 1, 24 (2013).
Pichardo, S., Sin, V. W. & Hynynen, K. Multi-frequency characterization of the speed of sound and attenuation coefficient for longitudinal transmission of freshly excised human skulls. Phys. Med. Biol. 56, 219–250 (2011).
Nyborg, W. L. Solutions of the bio-heat transfer equation. Phys. Med. Biol. 33, 785–792 (1988).
Zeineddine, H. A. et al. Subarachnoid blood clearance and aneurysmal subarachnoid hemorrhage outcomes: a retrospective review. Neurocrit. Care 39, 172–179 (2023).
Keep, R. F., Hua, Y. & Xi, G. Intracerebral haemorrhage: mechanisms of injury and therapeutic targets. Lancet Neurol. 11, 720–731 (2012).
Smyth, L. C. D. et al. Identification of direct connections between the dura and the brain. Nature 627, 165–173 (2024).
Plog, B. A. et al. A route for cerebrospinal fluid flow through leptomeningeal arterial–venous overlaps enables macromolecule and fluid shunting. Nat. Neurosci. 28, 1436–1445 (2025).
Ohashi, S. N. et al. Role of inflammatory processes in hemorrhagic stroke. Stroke 54, 605–619 (2023).
Escartin, C. et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 24, 312–325 (2021).
Wilhelmsson, U. et al. Redefining the concept of reactive astrocytes as cells that remain within their unique domains upon reaction to injury. Proc. Natl Acad. Sci. USA 103, 17513–17518 (2006).
Paolicelli, R. C. et al. Microglia states and nomenclature: a field at its crossroads. Neuron 110, 3458–3483 (2022).
Deczkowska, A. et al. Disease-associated microglia: a universal immune sensor of neurodegeneration. Cell 173, 1073–1081 (2018).
Guo, S., Wang, H. & Yin, Y. Microglia polarization from M1 to M2 in neurodegenerative diseases. Front. Aging Neurosci. 14, 815347 (2022).
Zheng, Z. V. & Wong, K. C. G. Microglial activation and polarization after subarachnoid hemorrhage. Neurosciences 6, 1 (2019).
Lier, J., Streit, W. J. & Bechmann, I. Beyond activation: characterizing microglial functional phenotypes. Cells 10, 2236 (2021).
Lalancette-Hébert, M. et al. Galectin-3 is required for resident microglia activation and proliferation in response to ischemic injury. J. Neurosci. 32, 10383–10395 (2012).
Gómez Morillas, A., Besson, V. C. & Lerouet, D. Microglia and neuroinflammation: what place for P2RY12?. Int. J. Mol. Sci. 22, 1636 (2021).
Treeby, B. E. & Cox, B. T. k-Wave: MATLAB toolbox for the simulation and reconstruction of photoacoustic wave fields. J. Biomed. Opt. 15, 021314 (2010).
Hersh, D. S. et al. Pulsed ultrasound expands the extracellular and perivascular spaces of the brain. Brain Res. 1646, 543–550 (2016).
Gnanasambandam, R. et al. GsMTx4: mechanism of inhibiting mechanosensitive ion channels. Biophys. J. 112, 31–45 (2017).
Suchyna, T. M. et al. Bilayer-dependent inhibition of mechanosensitive channels by neuroactive peptide enantiomers. Nature 430, 235–240 (2004).
Jin, H. et al. Increased CSF drainage by non-invasive manipulation of cervical lymphatics. Nature 643, 755–767 (2025).
Baek, H. et al. Clinical intervention using focused ultrasound (FUS) stimulation of the brain in diverse neurological disorders. Front. Neurol. 13, 880814 (2022).
Martin, E. et al. ITRUSST consensus on standardised reporting for transcranial ultrasound stimulation. Brain Stimul. 17, 607–615 (2024).
Attali, D. et al. Three-layer model with absorption for conservative estimation of the maximum acoustic transmission coefficient through the human skull for transcranial ultrasound stimulation. Brain Stimul. 16, 48–55 (2023).
Ye, D. et al. Mechanically manipulating glymphatic transport by ultrasound combined with microbubbles. Proc. Natl Acad. Sci. USA 120, e2212933120 (2023).
Holstein-Rønsbo, S. et al. Glymphatic influx and clearance are accelerated by neurovascular coupling. Nat. Neurosci. 26, 1042–1053 (2023).
Jiang-Xie, L.-F. et al. Neuronal dynamics direct cerebrospinal fluid perfusion and brain clearance. Nature 627, 157–164 (2024).
Chong, P. L. H., Garic, D., Shen, M. D., Lundgaard, I. & Schwichtenberg, A. J. Sleep, cerebrospinal fluid, and the glymphatic system: a systematic review. Sleep. Med Rev. 61, 101572 (2022).
Simon, M. J. & Iliff, J. J. Regulation of cerebrospinal fluid (CSF) flow in neurodegenerative, neurovascular and neuroinflammatory disease. Biochim. Biophys. Acta 1862, 442–451 (2016).
Hussain, R. et al. Potentiating glymphatic drainage minimizes post-traumatic cerebral oedema. Nature 623, 992–1000 (2023).
Seo, J.-S., Mantas, I., Svenningsson, P. & Greengard, P. Ependymal cells–CSF flow regulates stress-induced depression. Mol. Psychiatry 26, 7308–7315 (2021).
Kamp, M. A. et al. Evaluation of a murine single-blood-injection SAH model. PLoS ONE 9, e114946 (2014).
Rynkowski, M. A. et al. A mouse model of intracerebral hemorrhage using autologous blood infusion. Nat. Protoc. 3, 122–128 (2008).
Kiani Shabestari, S. et al. Absence of microglia promotes diverse pathologies and early lethality in Alzheimer’s disease mice. Cell Rep. 39, 110961 (2022).
Chen, W. et al. Modified behavioural tests to detect white matter injury- induced motor deficits after intracerebral haemorrhage in mice. Sci. Rep. 9, 16958 (2019).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 (2022).
Gong, C. et al. SAW: an efficient and accurate data analysis workflow for Stereo-seq spatial transcriptomics. GigaByte 2024, gigabyte111 (2024).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Fang, S. et al. Stereopy: modeling comparative and spatiotemporal cellular heterogeneity via multi-sample spatial transcriptomics. Nat. Commun. 16, 3741 (2025).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Cheng, M. et al. A cellular resolution spatial transcriptomic landscape of the medial structures in postnatal mouse brain. Front. Cell Dev. Biol. 10, 878346 (2022).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Palla, G. et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods 19, 171–178 (2022).
Hasgall, P. A., Neufeld, E., Gosselin, M. C., Klingenböck, A. & Kuster N. IT’IS database for thermal and electromagnetic parameters of biological tissues. ScienceOpen https://www.scienceopen.com/document?vid=a95fbaa4-efd8-429a-a59e-5e208fea2e45 (2012).
Ellis, P. D. The Essential Guide to Effect Sizes (Cambridge University Press, 2012).
Azadian, M. M. et al. Ultrasonic cerebrospinal fluid clearance improves outcomes in hemorrhagic brain injury models. Gene Expression Omnibus https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE296613 (2025).
Fukuda, A. M. & Badaut, J. Aquaporin 4: a player in cerebral edema and neuroinflammation. J. Neuroinflammation 9, 279 (2012).
Acknowledgements
We would like to thank J. J. Heit, M. Matrongolo, J. Foiret, Y. Guo, K. Ferrara, J. Dahl and K. Butts Pauly for a variety of useful discussions. We would like to thank R. Watkins for assistance with ultrasound hardware fabrication and characterization. We would like to thank the whole laboratory of R.D.A. and its alumni for helpful discussions and guidance for these experiments. We are grateful to K. Aschheim for support, insightful input and attentive editorial oversight. This study was supported by a Seed Grant from the Stanford Wu Tsai Neurosciences Institute (R.D.A.), NIH BRAIN Initiative (NIH/NIMH RF1MH114252 and NIH/NINDS UG3NS114438 to R.D.A.), NIH HEAL Initiative (NIH/NINDS UG3NS115637 to R.D.A.), NIH/NINDS R01NS126761 (P.M.G.), Focused Ultrasound Foundation (High Risk Award to R.D.A.), an anonymous donor to the Stanford SOM Radiology Department (R.D.A.), a Ford Foundation Predoctoral Fellowship (awarded to M.M.A.), an NSF Graduate Research Fellowship (awarded to B.J.Y.), a Hydrocephalus Association Innovator Award (R.M.F.), the Shurl and Kay Curci Foundation (R.M.F.), a Complete Genomics Spatial Biology Grant (R.D.A. and R.M.F.) and NIH 5-T32 GM141828-02 (A.R.).
Author information
Authors and Affiliations
Contributions
Conceptualization: M.M.A. and R.D.A. Methodology: M.M.A., S.K.S., A.R., P.J.M., N.M., E.M., Y.X., B.J.Y., P.M.G., R.M.F. and R.D.A. Investigation: M.M.A., S.K.S., A.R. and P.J.M. Visualization: M.M.A., A.R., P.J.M., R.M.F. and R.D.A. Funding acquisition: R.D.A., R.M.F. and P.M.G. Project administration: R.D.A. Supervision: R.D.A. Writing, original draft: M.M.A. and R.D.A. Writing, review and editing: M.M.A., P.J.M., R.M.F. and R.D.A.
Corresponding author
Ethics declarations
Competing interests
R.D.A. has received consulting fees from Cordance Medical and Lumos Labs and grant funding from AbbVie. All other authors declare no conflicts of interest.
Peer review
Peer review information
Nature Biotechnology thanks Max Tischfield and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Focused ultrasound parameter optimization for UDC.
a. Timeline of FUS parameters assessment experiment. b. (Left) Schematic of 650 kHz and 250 kHz transducer. Each transducer used was f = 1.0 with 30 mm (650 kHz) or 70 or 100 mm (250 kHz) aperture. (Right) Fluorescence intensity after pulsed ultrasound application (0.45 MPa estimated peak in situ negative pressure, 25% duty cycle, 5 Hz PRF, 10 min) at the indicated ultrasound frequency or sham (CNTRL) with quantification of mean fluorescence intensity after intracisternal injection of a 2 kDa tracer dye at 60 min post-injection. c. (Left) Representative coronal brain slice and quantification of mean fluorescence intensity after intracisternal injection of a 2 kDa tracer dye at 60 min post-injection. (Right) Given the inflection at 0.45 MPa estimated in situ peak negative pressure with saturation of the effect at higher pressures, this pressure was chosen for subsequent analysis. b. Data presented as box plots that show minima, maxima, interquartile range (box bounds) and median (black line). Kruskal-Wallis with post-hoc Dunn’s multiple comparisons test. n = 4 mice at each frequency; two-tailed Mann-Whitney U-test; Cntrl vs FUS/250 kHz: adjusted p = 0.0071. **: p ≤ 0.01. c. Data presented as mean ± S.D.; n = 3 mice at each pressure.
Extended Data Fig. 2 Focused ultrasound induced thermal effects.
a. (Left) Simulated heat accumulation for a single cycle of UDC, without consideration of thermoregulation due to brain perfusion. (Right) Experimentally measured temperature via a thermocouple placed intracranially before and after UDC application (thermocouple removed for UDC application with <5 sec time to replace the probe). b,c, kWave 3D heating simulation outputs in coronal (b) and sagittal (c) plane. Outputs include the resulting acoustic pressure amplitude (top left), volume rate of heat deposition (top center), temperature after the 50 ms burst of FUS (top right), and temperature after cooling 150 ms before the next ultrasound burst (bottom left). The remaining outputs show voxels above the CEM43 temperature threshold (bottom middle) and ablated tissue (bottom right), which were entirely zero for these analyses. a. Data presented as box plots that show minima, maxima, interquartile range (box bounds) and median (black line); n = 3 mice per group, two-tailed Mann-Whitney U-test, p = 0.200.
Extended Data Fig. 3 A pharmacologic benchmark mechanosensitive ion channel agonist is unsafe in ICH and fails to clear the hematoma.
a. Timeline of intracerebral hemorrhage (ICH) experiment with a mechanosensitive ion channel agonist (Yoda-1) administered intraperitoneally per dosing of prior protocols22. b. Increased survival in ICH with the UDC intervention (83.3%) compared to sham (50%), but decreased survival with Yoda-1 (16.7%), χ2(2) = 13.40, p < 0.05. A separate cohort with intracisternal Yoda-1 administration showed similarly low survival (1/5 survival at 2 days; 0/5 survival at 3 days). c. Administration of Yoda-1 did not reduce blood product presence in contrast to UDC, as assessed by hematoma volumes across treatment groups. Data presented as box plots that show minima, maxima, interquartile range (box bounds) and median (black line); healthy control data are presented for reference. Kruskal-Wallis with post-hoc Dunn’s multiple comparisons test. Kruskal-Wallis with post-hoc Dunn’s multiple comparisons test. UDC, Sham, and GsMTx4 data repeated from Fig. 5. *: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001. Initial n = 6-8 mice per group.
Extended Data Fig. 4 Schematic for the proposed hypothetical model of the mechanisms underlying the efficacy of UDC.
In hemorrhagic stroke, aquaporin expression and disease-associated microglial states are promoted (Fig. 4). Disease-associated microglia inhibit the polarization of aquaporins to astrocytic endfeet75. UDC acts principally via mechanosensitive ion channel activation (Fig. 6) and potentially also via direct mechanical fluid compartmental intermixing (Extended Data Fig. 1)25. Via mechanosensitive ion channel activation, UDC shifts microglia from the disease-associated to the homeostatic state (Figs. 4, 5, 6), in accord with prior literature18,19,20. This microglial phenotypic shift yields increased RBC phagocytosis (Fig. 5) and disinhibition of astrocytic endfeet aquaporin polarization (Figs. 4, 5). The combination of these effects (increased mechanical fluid mixing, aquaporin polarization, and debris phagocytosis) allows for clearance of brain injury debris via the meningeal lymphatics to the cervical lymph nodes (Fig. 1) and possibly – though less directly supported in our presented data – via arachnoid villi to the dural venous sinuses.
Supplementary information
Supplementary Video 1
Microglial phagocytosis of red blood cells in reference to 3D histology for quantification of Fig. 5f.
Source data
Source Data Figs. 1–3, 5 and 6 and Extended Data Figs. 1–3
Raw source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Azadian, M.M., Kiani Shabestari, S., Rajan, A. et al. Clearance of intracranial debris by ultrasound reduces inflammation and improves outcomes in hemorrhagic stroke models. Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02866-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41587-025-02866-8