Abstract
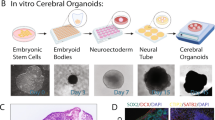
Profiling protein abundance and dynamics at single-cell resolution in complex human tissues is challenging. Given the discordance between transcript and protein abundance observed in studies of the human cerebral cortex, we developed an optimized workflow that combines label-free single-cell mass spectrometry with precise sample preparation to resolve quantitative proteomes of individual cells from the developing human brain. Our method achieves deep proteomic coverage (~800 proteins per cell) even in small immature prenatal human neurons (diameter ~7–10 μm, ~50 pg protein), capturing major brain cell types and enabling proteome-wide characterization at single-cell resolution. We document extensive transcriptome–proteome discordance across cell types, particularly in genes associated with neurodevelopmental disorders. Proteins exhibit markedly higher cell-type specificity than their mRNA counterparts, underscoring the importance of proteomic-level analysis. By reconstructing developmental trajectories from radial glia to excitatory neurons at the proteomic level, we identify dynamic, stage-specific protein co-expression modules and pinpoint the intermediate progenitor-to-neuron transition as a genetically vulnerable phase associated with autism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Processed single-cell proteome and single-cell transcriptome data are available at an interactive portal (https://cell.ucsf.edu/SCProteome/). Single-cell proteomics raw data have been deposited to the ProteomeXchange Consortium (PXD071075)91. The single-cell transcriptome data are available in the Gene Expression Omnibus repository (GSE310125)92. The reference single-cell RNA-seq atlas of the developing human brain39 is accessible at https://cell.ucsf.edu/snMultiome/. Bulk proteome data were provided in Supplementary Tables 2 and 3. Bulk transcriptome data were sourced from the BrainSpan atlas (https://www.brainspan.org/static/download.html). The list of short-lived proteins was obtained from a previous study62. Synaptic genes were identified using SynaptomeDB42, and the SFARI gene list was downloaded from the SFARI Gene database (https://gene.sfari.org/database/human-gene/). Data for mutations associated with autism and NDDs were sourced from a previous study66. Protein pLI scores were obtained from gnomAD93, and evolutionary rates were retrieved from the Ensembl genome browser (v99) through biomart94. The polysome TrIP-seq data were retrieved from previous study57. The list of human transcription factors is available at https://humantfs.ccbr.utoronto.ca/download/v_1.01/TF_names_v_1.01.txt.
Code availability
The code used for data analysis is available on GitHub (https://github.com/SingleCellProteomics/Brain)95.
References
Gry, M. et al. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics 10, 365 (2009).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016).
Jiang, L. et al. A quantitative proteome map of the human body. Cell 183, 269–283 (2020).
Wang, D. et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 15, e8503 (2019).
Sharma, K. et al. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 18, 1819–1831 (2015).
Takemon, Y. et al. Proteomic and transcriptomic profiling reveal different aspects of aging in the kidney. eLife 10, e62585 (2021).
Ghazalpour, A. et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 7, e1001393 (2011).
Tracey, L. J., An, Y. & Justice, M. J. CyTOF: an emerging technology for single-cell proteomics in the mouse. Curr. Protoc. 1, e118 (2021).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Aebersold, R. & Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016).
Anand, S., Samuel, M., Ang, C.-S., Keerthikumar, S. & Mathivanan, S. Label-based and label-free strategies for protein quantitation. Methods Mol. Biol. 1549, 31–43 (2017).
Virant-Klun, I., Leicht, S., Hughes, C. & Krijgsveld, J. Identification of maturation-specific proteins by single-cell proteomics of human oocytes. Mol. Cell. Proteomics 15, 2616–2627 (2016).
Budnik, B., Levy, E., Harmange, G. & Slavov, N. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 19, 161 (2018).
Wang, Z., Liu, P.-K. & Li, L. A tutorial review of labeling methods in mass spectrometry-based quantitative proteomics. ACS Meas. Sci. Au 4, 315–337 (2024).
Brenes, A., Hukelmann, J., Bensaddek, D. & Lamond, A. I. Multibatch TMT reveals false positives, batch effects and missing values. Mol. Cell. Proteomics 18, 1967–1980 (2019).
Phua, S.-X., Lim, K.-P. & Goh, W. W.-B. Perspectives for better batch effect correction in mass-spectrometry-based proteomics. Comput. Struct. Biotechnol. J. 20, 4369–4375 (2022).
Guzman, U. H. et al. Ultra-fast label-free quantification and comprehensive proteome coverage with narrow-window data-independent acquisition. Nat. Biotechnol. 42, 1855–1866 (2024).
Brunner, A. et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. Mol. Syst. Biol. 18, e10798 (2022).
Schoof, E. M. et al. Quantitative single-cell proteomics as a tool to characterize cellular hierarchies. Nat. Commun. 12, 3341 (2021).
Furtwängler, B. et al. Mapping early human blood cell differentiation using single-cell proteomics and transcriptomics. Science 390, eadr8785 (2025).
Lombard-Banek, C., Reddy, S., Moody, S. A. & Nemes, P. Label-free quantification of proteins in single embryonic cells with neural fate in the cleavage-stage frog (Xenopus laevis) embryo using capillary electrophoresis electrospray ionization high-resolution mass spectrometry (CE-ESI-HRMS). Mol. Cell. Proteomics 15, 2756–2768 (2016).
Bubis, J. A. et al. Challenging the Astral mass analyzer to quantify up to 5,300 proteins per single cell at unseen accuracy to uncover cellular heterogeneity. Nat. Methods 22, 510–519 (2025).
Mun, D.-G. et al. Diversity of post-translational modifications and cell signaling revealed by single cell and single organelle mass spectrometry. Commun. Biol. 7, 884 (2024).
Mun, D.-G. et al. Optimizing single cell proteomics using trapped ion mobility spectrometry for label-free experiments. Analyst 148, 3466–3475 (2023).
Sanchez-Avila, X. et al. Easy and accessible workflow for label-free single-cell proteomics. J. Am. Soc. Mass Spectrom. 34, 2374–2380 (2023).
Fang, W. et al. A rapid and sensitive single-cell proteomic method based on fast liquid-chromatography separation, retention time prediction and MS1-only acquisition. Anal. Chim. Acta 1251, 341038 (2023).
Guo, Y. et al. Single-cell quantitative proteomic analysis of human oocyte maturation revealed high heterogeneity in in vitro-matured oocytes. Mol. Cell. Proteomics 21, 100267 (2022).
Tsai, C.-F. et al. Surfactant-assisted one-pot sample preparation for label-free single-cell proteomics. Commun. Biol. 4, 265 (2021).
Cong, Y. et al. Improved single-cell proteome coverage using narrow-bore packed nanoLC columns and ultrasensitive mass spectrometry. Anal. Chem. 92, 2665–2671 (2020).
Lombard-Banek, C., Moody, S. A., Manzini, M. C. & Nemes, P. Microsampling capillary electrophoresis mass spectrometry enables single-cell proteomics in complex tissues: developing cell clones in live Xenopus laevis and zebrafish embryos. Anal. Chem. 91, 4797–4805 (2019).
Dou, M. et al. High-throughput single cell proteomics enabled by multiplex isobaric labeling in a nanodroplet sample preparation platform. Anal. Chem. 91, 13119–13127 (2019).
Ai, L. et al. Single-cell proteomics reveals specific cellular subtypes in cardiomyocytes derived from human iPSCs and adult hearts. Mol. Cell. Proteomics 24, 100910 (2025).
Kreimer, S. et al. High-throughput single-cell proteomic analysis of organ-derived heterogeneous cell populations by nanoflow dual-trap single-column liquid chromatography. Anal. Chem. 95, 9145–9150 (2023).
Rosenberger, F. A. et al. Spatial single-cell mass spectrometry defines zonation of the hepatocyte proteome. Nat. Methods 20, 1530–1536 (2023).
Matzinger, M., Müller, E., Dürnberger, G., Pichler, P. & Mechtler, K. Robust and easy-to-use one-pot workflow for label-free single-cell proteomics. Anal. Chem. 95, 4435–4445 (2023).
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020).
Ye, Z. et al. Enhanced sensitivity and scalability with a Chip-Tip workflow enables deep single-cell proteomics. Nat. Methods 22, 499–509 (2025).
Géminard, C., de Gassart, A. & Vidal, M. Reticulocyte maturation: mitoptosis and exosome release. Biocell 26, 205–215 (2002).
Wang, L. et al. Molecular and cellular dynamics of the developing human neocortex. Nature 647, 169–178 (2025).
Shi, Y. et al. Mouse and human share conserved transcriptional programs for interneuron development. Science 374, eabj6641 (2021).
Wang, L. et al. A cross-species proteomic map reveals neoteny of human synapse development. Nature 622, 112–119 (2023).
Pirooznia, M. et al. SynaptomeDB: an ontology-based knowledgebase for synaptic genes. Bioinformatics 28, 897–899 (2012).
Miller, J. A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014).
Perez, J. D. et al. Subcellular sequencing of single neurons reveals the dendritic transcriptome of GABAergic interneurons. eLife 10, e63092 (2021).
Li, L. T., Jiang, G., Chen, Q. & Zheng, J. N. Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol. Med. Rep. 11, 1566–1572 (2015).
Heidebrecht, H. J., Buck, F., Haas, K., Wacker, H. H. & Parwaresch, R. Monoclonal antibodies Ki-S3 and Ki-S5 yield new data on the ‘Ki-67’ proteins. Cell Prolif. 29, 413–425 (1996).
Tani, H. et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 22, 947–956 (2012).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Hevner, R. F. et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29, 353–366 (2001).
Raju, C. S. et al. Secretagogin is expressed by developing neocortical GABAergic neurons in humans but not mice and increases neurite arbor size and complexity. Cereb. Cortex 28, 1946–1958 (2017).
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Drivas, T. G. et al. A second cohort of CHD3 patients expands the molecular mechanisms known to cause Snijders Blok-Campeau syndrome. Eur. J. Hum. Genet. 28, 1422–1431 (2020).
Washbourne, P. Synapse assembly and neurodevelopmental disorders. Neuropsychopharmacology 40, 4–15 (2015).
Sapir, T., Frotscher, M., Levy, T., Mandelkow, E.-M. & Reiner, O. Tau’s role in the developing brain: implications for intellectual disability. Hum. Mol. Genet. 21, 1681–1692 (2012).
Fiock, K. L., Smalley, M. E., Crary, J. F., Pasca, A. M. & Hefti, M. M. Increased tau expression correlates with neuronal maturation in the developing human cerebral cortex. eNeuro 7, ENEURO.0058–20.2020 (2020).
Wang, J. et al. Benchmarking informatics workflows for data-independent acquisition single-cell proteomics. Nat. Commun. 16, 10276 (2025).
Blair, J. D., Hockemeyer, D., Doudna, J. A., Bateup, H. S. & Floor, S. N. Widespread translational remodeling during human neuronal differentiation. Cell Rep. 21, 2005–2016 (2017).
Thumuluri, V. et al. NetSolP: predicting protein solubility in Escherichia coli using language models. Bioinformatics 38, 941–946 (2022).
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Nagore, L. I. et al. Purification and characterization of transcription factors. Mass Spectrom. Rev. 32, 386–398 (2013).
Mauger, O., Lemoine, F. & Scheiffele, P. Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron 92, 1266–1278 (2016).
Li, J. et al. Proteome-wide mapping of short-lived proteins in human cells. Mol. Cell 81, 4722–4735 (2021).
Blake, J. A. et al. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 42, D810–D817 (2014).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Abrahams, B. S. et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4, 36 (2013).
Zhou, X. et al. Integrating de novo and inherited variants in 42,607 autism cases identifies mutations in new moderate-risk genes. Nat. Genet. 54, 1305–1319 (2022).
Park, C. Y. et al. Genome-wide landscape of RNA-binding protein target site dysregulation reveals a major impact on psychiatric disorder risk. Nat. Genet. 53, 166–173 (2021).
Yanai, I. et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21, 650–659 (2005).
Webster, E. et al. De novo PHIP-predicted deleterious variants are associated with developmental delay, intellectual disability, obesity, and dysmorphic features. Cold Spring Harb. Mol. Case Stud. 2, a001172 (2016).
Kampmeier, A. et al. PHIP-associated Chung-Jansen syndrome: report of 23 new individuals. Front. Cell Dev. Biol. 10, 1020609 (2022).
Mattioli, F. et al. De novo frameshift variants in the neuronal splicing factor NOVA2 result in a common C-terminal extension and cause a severe form of neurodevelopmental disorder. Am. J. Hum. Genet. 106, 438–452 (2020).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Melliou, S. et al. Regionally defined proteomic profiles of human cerebral tissue and organoids reveal conserved molecular modules of neurodevelopment. Cell Rep. 39, 110846 (2022).
Machol, K. et al. Expanding the spectrum of BAF-related disorders: de novo variants in SMARCC2 cause a syndrome with intellectual disability and developmental delay. Am. J. Hum. Genet. 104, 164–178 (2019).
Schanze, I. et al. NFIB haploinsufficiency is associated with intellectual disability and macrocephaly. Am. J. Hum. Genet. 103, 752–768 (2018).
Beck, D. B. et al. A recurrent de novo CTBP1 mutation is associated with developmental delay, hypotonia, ataxia, and tooth enamel defects. Neurogenetics 17, 173–178 (2016).
Gingold, H. et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 (2014).
Raj, A. & van Oudenaarden, A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226 (2008).
Kelleher, R. J. & Bear, M. F. The autistic neuron: troubled translation? Cell 135, 401–406 (2008).
Darnell, J. C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011).
Hooshmandi, M., Wong, C. & Khoutorsky, A. Dysregulation of translational control signaling in autism spectrum disorders. Cell. Signal. 75, 109746 (2020).
Sanes, J. R. & Zipursky, S. L. Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell 181, 536–556 (2020).
Díaz-Villanueva, J. F., Díaz-Molina, R. & García-González, V. Protein folding and mechanisms of proteostasis. Int. J. Mol. Sci. 16, 17193–17230 (2015).
Clapier, C. R., Iwasa, J., Cairns, B. R. & Peterson, C. L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017).
Ting, L., Rad, R., Gygi, S. P. & Haas, W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 (2011).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
PRIDE. Single-cell proteomic landscape of the developing human brain. www.ebi.ac.uk/pride/archive/projects/PXD071075/ (2025).
Gene Expression Omnibus. Single-cell transcriptomic profiling of the developing human brain (GW 13, 15, and 19). www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE310125 (2025).
Chen, S. et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 625, 92–100 (2024).
Kinsella, R. J. et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011, bar030 (2011).
Tianzhi, W. Single-cell proteomic landscape in the developing human brain. GitHub github.com/SingleCellProteomics/Brain (2025).
Acknowledgements
This work was supported by the SFARI grant (AN-AR-Gene Therapies-01010017 to J.L., A.R.K. and M.S.).
Author information
Authors and Affiliations
Contributions
J.L., L.J. and T.W. designed the study. L.W., T.M. and A.R.K. helped with sample collection. T.W., L.J., T.M., R.J., T.T., J.L. and M.S. were responsible for single-cell proteomic data production. L.W. was responsible for generating single-cell RNA-seq data. T.M. documented immunostaining validation. T.W. and T.M. conducted the immunostaining data analysis. T.W. and J.L. conducted the single-cell data analysis. J.L., A.R.K. and M.S. were responsible for funding. J.L., T.W., L.W., L.J. and A.R.K. were involved in writing the original draft and performing the revision. All authors contributed to the review and editing of the paper.
Corresponding authors
Ethics declarations
Competing interests
A.R.K. is a cofounder, consultant and director of Neurona Therapeutics. J.L. is a cofounder of SensOmics and serves on its scientific advisory board. M.S. is a cofounder of Personalis, SensOmics, Qbio, January AI, Filtricine, Protos and NiMo, and serves on the scientific advisory boards of Personalis, SensOmics, Qbio, January AI, Filtricine, Protos, NiMo and Genapsys. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Bogdan Budnik and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note (quantitative data analysis) and Supplementary Figs. 1–21.
Supplementary Tables
Supplementary Tables 1–12.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, T., Jiang, L., Mukhtar, T. et al. Single-cell proteomic landscape of the developing human brain. Nat Biotechnol (2026). https://doi.org/10.1038/s41587-025-02980-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41587-025-02980-7