Abstract
Transcriptome-wide association analysis is a powerful approach to studying the genetic architecture of complex traits. A key component of this approach is to build a model to impute gene expression levels from genotypes by using samples with matched genotypes and gene expression data in a given tissue. However, it is challenging to develop robust and accurate imputation models with a limited sample size for any single tissue. Here, we first introduce a multi-task learning method to jointly impute gene expression in 44 human tissues. Compared with single-tissue methods, our approach achieved an average of 39% improvement in imputation accuracy and generated effective imputation models for an average of 120% more genes. We describe a summary-statistic-based testing framework that combines multiple single-tissue associations into a powerful metric to quantify the overall gene–trait association. We applied our method, called UTMOST (unified test for molecular signatures), to multiple genome-wide-association results and demonstrate its advantages over single-tissue strategies.
Similar content being viewed by others
Main
Genome-wide association studies (GWAS) have successfully identified numerous single-nucleotide polymorphisms (SNPs) associated with complex human traits and diseases. Despite these successes, important problems remain in statistical power and biological interpretation of GWAS results2. In particular, the complex architecture of linkage disequilibrium and context-dependent regulatory machinery in the genome hinder the ability to accurately identify disease genes from GWAS, thereby raising challenges in downstream functional validation and therapeutics development. Recently, large-scale consortia, such as the Genotype-Tissue Expression (GTEx) project3,4, have generated matched genotype and expression data for various human tissues. These rich datasets have provided great insights into the mechanisms of cross-tissue transcriptional regulation and accelerated discoveries for expression quantitative trait loci (eQTL)4,5,6,7. In addition, integrating eQTL information in genetic association analysis has become an effective way to bridge SNPs, genes, and complex traits. Many methods have been developed to co-localize eQTL with loci identified in GWAS to identify candidate risk genes for complex traits8,9,10,11,12,13. Two recent studies have addressed this issue through an innovative approach that is sometimes referred to as transcriptome-wide association analysis. First, on the basis of an externally trained imputation model, gene expression is imputed by using genotype information in GWAS samples. Next, gene-level association is assessed between imputed gene expression and the trait of interest14,15. These methods have gained popularity in the past two years, owing to their capability to effectively use signals from multiple eQTL with moderate effects and to reduce the effect of reverse causality in expression–trait association analysis. The applications of these methods have led to novel insights into the genetic basis of many diseases and traits16,17,18.
Despite these successes, existing methods have several limitations. First, owing to the tissue-dependent nature of transcription regulation, existing methods train separate imputation models for different tissues. This practice ignores the similarity in transcription regulation across tissues, thereby limiting the effective sample sizes for tissues that are difficult to acquire. Second, a hypothesis-free search across genes and tissues increases the burden of multiple testing and thus reduces statistical power. Pinpointing a subset of tissues according to prior knowledge may resolve this issue to some extent. However, for many complex traits, biologically relevant tissues are unknown. Further, reports have shown that eQTL with large effects tend to regulate gene expression in multiple tissues4. Genetic correlation analysis has also suggested substantial sharing of local expression regulation across tissues19. This would inevitably result in statistically significant associations in tissues irrelevant to the trait of interest, a phenomenon that has been extensively discussed recently20. Jointly analyzing data from multiple genetically correlated tissues has the potential to resolve these issues. Multi-trait analysis has been demonstrated to improve the accuracy of genetic risk prediction21,22,23. Multi-tissue modeling has also been shown to improve the statistical power in eQTL discovery24,25,26,27 and gene network studies28. In this work, we demonstrate that a cross-tissue strategy can also improve transcriptome-wide association analysis.
We introduce UTMOST, a principled method to perform cross-tissue expression imputation and gene-level association analysis. We demonstrate its performance through internal and external imputation validation, simulation studies, analyses of 50 complex traits, a case study on low-density-lipoprotein cholesterol (LDL-C), and a multi-stage association study for late-onset Alzheimer’s disease (LOAD). We show that UTMOST substantially improves the accuracy of expression imputation in all available tissues. In the downstream association analysis, UTMOST provides a powerful metric that summarizes gene-level associations across tissues and can be extended to integrate various molecular phenotypes.
Results
Model overview
The UTMOST framework consists of three main stages (Fig. 1). First, for each gene in the genome, we train a cross-tissue expression imputation model by using the genotype information and matched expression data from 44 tissues in GTEx. Next, we test associations between the trait of interest and imputed expression in each tissue. Finally, a cross-tissue test is performed for each gene to summarize single-tissue association statistics into a powerful metric that quantifies the overall gene–trait association. Here, we briefly introduce the UTMOST framework. All the statistical details are discussed in the Methods.
We formulate cross-tissue expression imputation as a penalized multivariate regression problem:
where N, M, and P denote the sample size in the training data, the number of SNPs in the imputation model, and the total number of tissues, respectively. Because only a subset of tissues was collected from each individual, expression data in matrix Y were incomplete, and sample sizes for different tissues were unbalanced. We estimate B by minimizing the squared loss function with a lasso penalty on the columns (within-tissue effects) and a group-lasso penalty on the rows (cross-tissue effects) (Methods).
where Yi, Xi, and Ni denote the observed expressions, genotypes, and sample size of the ith tissue, respectively. Parameters λ1 and λ2 are tuned through cross-validation. Our cross-tissue imputation model does not assume eQTL to have the same effect direction across tissues. Instead, UTMOST uses a group-lasso29 penalty term to encourage the presence of cross-tissue eQTL and improve the estimation of their effects.
In the second stage, we test the associations between the trait of interest and imputed gene expression in each tissue. We denote imputed gene expression in the ith tissue as \(E_i = X_i\hat B_{ \cdot i}\) and test associations via a univariate regression model:
where αi is the intercept, γi is the effect size of gene expression, and δi denotes the residual. The z scores for gene–trait associations in the ith tissue can be denoted as:
where \(\tilde Z\) denotes the SNP-trait z scores, and Γi is a diagonal matrix whose jth diagonal element denotes the ratio between the standard deviation of the jth SNP and that of imputed expression in the ith tissue (Methods). When there is no SNP–trait association, \(\tilde Z\) follows a multivariate normal distribution N(0,D), where D is the linkage-disequilibrium matrix for SNPs. The covariance matrix of Z = (Z1, Z2, …, ZP)T can be calculated as:
where \({\mathrm{\Lambda }} = (\hat B_{ \cdot 1}{\mathrm{\Gamma }}_1,\hat B_{ \cdot 2}{\mathrm{\Gamma }}_2, \ldots ,\hat B_{ \cdot P}{\mathrm{\Gamma }}_P)\).
Finally, we combine single-tissue gene–trait association results by using a generalized Berk–Jones (GBJ) test, which takes the covariance among single-tissue test statistics into account30. We note that this framework allows gene–trait associations to have different directions across tissues. Details on the GBJ statistic and P-value calculation are discussed in the Methods.
Cross-tissue expression imputation accuracy
We first evaluated the accuracy of cross-tissue expression imputation through fivefold cross-validation. We used an elastic net model (that is, the model used in PrediXcan14) trained in each tissue separately as the benchmark for prediction without leveraging cross-tissue information. We used squared Pearson correlation (R2) between the observed and predicted gene expression levels to quantify imputation accuracy. Cross-tissue imputation achieved higher imputation accuracy in all 44 tissues (Fig. 2a). On average, imputation accuracy was improved by 38.6% across tissues (Fig. 2b). The improvement was particularly high in tissues with low sample sizes in GTEx (N < 150; average of 47.4% improvement). Analysis based on Spearman correlation also showed consistent results (Supplementary Fig. 1). Next, we calculated the proportion of genes with increased imputation accuracy. In all 44 tissues, substantially more genes showed improved imputation performance (Supplementary Table 1). With a false discovery rate (FDR) cutoff of 0.05 as the significance threshold, our cross-tissue method achieved 120% more significantly predicted genes across tissues. Among tissues with low sample sizes, the improvement percentage rose even further to 175% (Fig. 2c). Furthermore, we compared our method with the Bayesian sparse linear mixed-effects model (BSLMM31), the imputation method used in TWAS15. Similarly, UTMOST achieved higher imputation accuracy in all 44 tissues (Supplementary Fig. 2). On average, the imputation accuracy improved by 20.3% across tissues.
a,b Compared with single-tissue elastic net, UTMOST showed substantially higher average increment in R2 across genes (a) and relative improvement (that, is percentage of increment in R2) in imputation accuracy (b). c, UTMOST identified more imputed genes, especially in tissues with smaller sample sizes in GTEx. Sample sizes of 44 GTEx tissues are listed in Supplementary Table 1, with predictability tested by F test with d.f. of 1 and n – 2. d,e, The imputation improvement in two specific examples in whole-blood tissue, where the shaded region represents the 95%-confidence band.
Next, we performed external validation, using two independent datasets. We first used our imputation model for whole blood in GTEx to predict gene expression levels in GEUVADIS lymphoblastoid cell lines (LCLs)32 (Methods). The imputation accuracy quantified as R2 showed substantial departure from the expected distribution under the null hypothesis (that is, expression and SNPs are independent), thus demonstrating the generalizability of cross-tissue imputation (Supplementary Figs. 3 and 4). Compared with single-tissue elastic net, cross-tissue imputation achieved significantly higher prediction accuracy in different quantiles (P = 3.43 × 10−7; Kolmogorov–Smirnov test), in agreement with our findings from cross-validation. Two examples of well-predicted genes are illustrated in Fig. 2d,e, showing improved concordance between observed (gene expression adjusted for potential confounding effects; Methods) and predicted expression values via cross-tissue imputation. Analysis on CommonMind consortium data33 showed similar results (Methods; Supplementary Figs. 5 and 6).
Cross-tissue association test
Another key advancement in the UTMOST framework is a novel gene-level association test that combines statistical evidence across multiple tissues. We performed simulation studies using samples from the Genetic Epidemiology Research Study on Adult Health and Aging (GERA; N = 12,637) to assess the association test’s type I error rate and statistical power in a variety of settings (Methods). We did not observe inflation in the type I error rate in two different simulation studies (Supplementary Tables 2 and 3). We observed a substantial improvement in statistical power of the multi-tissue joint test when gene expression in multiple tissues were causally related to the trait. The improvement was also consistent under different simulated genetic architectures (Fig. 3). When the trait was affected by expression in only one tissue, the statistical power of the joint test was comparable to that of a single-tissue test in the causal tissue. Compared with the naïve test, which combines results across tissues while applying an additional Bonferroni correction, our joint test was consistently more powerful (improvement ranged from 15.3% to 24.1%).
We compared the statistical power of UTMOST, a single-tissue association test, and a simple union of findings from single-tissue analysis with various disease architectures. Left/right panels represent the cases in which genes explain 1%/0.1% of trait variance in total (denoted as high/low phenotypic effects). Muscle is the only causal tissue in setting 1. Both muscle and skin are causal tissues in setting 2. All three tissues are causal in setting 3.
UTMOST identifies more associations in relevant tissues
To evaluate the performance of the single-tissue association test based on cross-tissue expression imputation, we applied UTMOST to the summary statistics from 50 GWAS (Ntotal ≈ 4.5 million without adjusting for sample overlap across studies; Supplementary Table 4) and compared the results with those of PrediXcan14 and TWAS15. To identify tissue types that are biologically relevant to these complex traits, we applied linkage-disequilibrium-score regression34 to these datasets and partitioned heritability by tissue-specific functional genome predicted by GenoSkyline-Plus annotations35. Tissue-trait relevance was ranked on the basis of enrichment P values (Methods). Compared with PrediXcan and TWAS, UTMOST identified substantially more associations in the most relevant tissue for each analyzed trait, showing 69.2% improvement compared with PrediXcan (P = 8.79 × 10−5; paired Wilcoxon rank test) and 188% improvement compared with TWAS (P = 7.39 × 10−8, Fig. 4). Such improvement was consistently observed across traits (Supplementary Table 5). In contrast, for other tissues, UTMOST identified similar numbers of genes and showed no significant difference compared with PrediXcan (P = 0.52). Comparing tissues that were most and least enriched for trait heritability, UTMOST identified significantly more associations in tissues strongly enriched for trait heritability than in tissues with the least enrichment (P = 0.016), whereas the contrast was not significant on the basis of PrediXcan (P = 0.192) or TWAS (P = 0.085). Finally, we applied the cross-tissue joint test to these traits and compared the number of significant genes with the combined results from 44 UTMOST single-tissue tests. The UTMOST joint test identified more associations than single-tissue tests in 43 out of 50 traits (P = 1.74 × 10−8; Wilcoxon rank test; Supplementary Fig. 7), showing improved statistical power in cross-tissue analysis.
Boxes on the left show the numbers of genes identified in all other tissues. Boxes on the right show the numbers of genes identified in the most relevant tissue for each trait. In each box, the two horizontal borders represent the upper and lower quartiles, and the solid line in the middle represents the median. The highest and lowest points indicate the maxima and minima. P values were calculated via one-sided paired Wilcoxon rank tests (N = 50).
Integrating external QTL resource
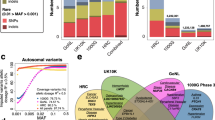
We applied UTMOST to the meta-analysis summary data of LDL-C from the Global Lipids Genetics Consortium (N = 173,082)36. Results based on four different analytical strategies, that is, single-tissue test using liver tissue in GTEx (N = 97), single-tissue test using liver eQTL from STARNET37 (N = 522), cross-tissue joint test combining 44 GTEx tissues, and cross-tissue joint test combining 44 GTEx tissues and the liver eQTL from STARNET, were compared. We identified 57, 58, 185, and 203 significant genes in the four sets of analyses, respectively (Fig. 5a).
a, Number of significant genes identified in four sets of analyses (z-score test for single-tissue and generalized Berk–Jones for cross-tissue test; Bonferroni-corrected thresholds were used, that is, 4.49 × 10-6, 8.39 × 10-6, 3.31 × 10-6, and 3.31 × 10-6). b, Associations at the SORT1 locus; values on the x axis were based on the transcription start site of each gene. The horizontal line indicates the Bonferroni-corrected genome-wide-significance threshold (N = 173,082, generalized Berk–Jones test).
Among the identified genes in the cross-tissue joint test of 44 GTEx tissues and STARNET liver, SORT1 had the most significant association (P = 3.4 × 10−15). SORT1 is known to causally mediate LDL-C levels, even though the GWAS association signal at this locus is clustered around CELSR2 (ref. 38,39). Of note, liver was not implicated as the relevant tissue for SORT1 in the association analysis, and moreover the association signal at SORT1 was completely absent in the single tissue test based on GTEx-liver, owing to its low imputation quality (FDR = 0.064). The limited sample size of liver tissue in GTEx (N = 97) restrained the imputation performance of SORT1 and consequently reduced the statistical power in the association test. In contrast, UTMOST successfully recovered the association signal at SORT1 (P = 3.4 × 10−15). Additionally, the UTMOST cross-tissue association test is flexible in incorporating external QTL resources along with GTEx data (Methods). Through integrating single-tissue associations in all 44 GTEx tissues and a large external liver dataset (STARNET; N = 522), we successfully recovered the association of SORT1 (Fig. 5b). Furthermore, we performed pairwise conditional analyses between SORT1 and other significant genes at the SORT1 locus, and we found that SORT1 remained statistically significant in all analyses, thus showing that its association signal is not shadowed by other genes (Supplementary Table 6). Further, when correlations between gene expression were moderate, SORT1 was more significant than all other tested genes in conditional analysis. Even when the correlation was substantial (for example, CELSR2 and PSRC1 both had correlation of 0.9 with SORT1 in STARNET), SORT1 remained statistically significant. We compared association based on STARNET only and found that SORT1 was not the top signal in the locus in single-tissue analysis, and the cross-tissue approach did not increase the false-positive rate (Supplementary Note). These results suggest that integrative analysis of transcriptomic data from multiple tissues and multiple QTL resources can effectively increase statistical power in gene-level association mapping. UTMOST is a flexible framework and is not limited to GTEx tissues only. Integrating relevant external QTL studies via UTMOST may further improve downstream association analysis.
UTMOST identifies novel risk genes for Alzheimer’s disease
Finally, to demonstrate UTMOST’s effectiveness in real association studies, we performed a multi-stage gene-level association study for LOAD. In the discovery stage, we applied UTMOST to the stage I GWAS summary statistics from the International Genomics of Alzheimer’s Project40 (IGAP; N = 54,162). A number of recent studies have suggested that functional DNA regions in liver and myeloid cells are strongly enriched for LOAD heritability35,41,42. In addition, alternative splicing has also been suggested to be a mechanism for many risk loci of LOAD43. Therefore, in addition to 44 tissues from GTEx, we also incorporated liver eQTL from STARNET and both eQTL and splicing (s)QTL data in three immune cell types (that is, CD14+ monocytes, CD16+ neutrophils, and naive CD4+ T cells) from the BLUEPRINT44 consortium in our analysis (Methods). Single-tissue association tests were performed and then combined by using the GBJ test. In total, our cross-tissue analysis identified 68 genome-wide-significant genes in the discovery stage (Supplementary Table 7 and Supplementary Fig. 8).
Next, we replicated our findings in two independent datasets: using GWAS summary statistics based on samples in the Alzheimer’s Disease Genetics Consortium (ADGC) that were not used in the IGAP stage I analysis (N = 7,050) and summary statistics from the GWAS by proxy45 (GWAX; N = 114,564). Despite the moderate sample size in the ADGC dataset and the ‘proxy’ LOAD phenotype based on family history in GWAX analysis, the replication rate was high (Supplementary Table 7). Out of 68 genes, 17 and 15 were successfully replicated under the Bonferroni-corrected significance threshold in ADGC and GWAX, respectively. The numbers of replicated genes rose to 41 and 30 under a relaxed P-value cutoff of 0.05. Twenty-two out of 68 genes had P values below 0.05 in both replication datasets. We then combined P values from all three analyses via Fisher’s method. A total of 69 genes, including 12 genes that were not significant in the discovery stage, reached genome-wide significance in the meta-analysis (Fig. 6 and Supplementary Table 7 and 8). These 69 genes were significantly enriched for seven gene ontology terms (Supplementary Table 9), and ‘very-low-density-lipoprotein particle’ was the most significant (adjusted P = 5.8 × 10−3).
Most significant genes are from previously identified LOAD risk loci40,46,47,48,49,50,51. These include the CR1 locus on chromosome 1, BIN1 locus on chromosome 2, HBEGF locus on chromosome 5, ZCWPW1 and EPHA1 loci on chromosome 7, CLU locus on chromosome 8, CELF1, MS4A6A, and PICALM loci on chromosome 11, and APOE region on chromosome 19. Among these loci, AGFG2 rather than ZCWPW1, the previously suggested index gene at this locus40, was significant in the meta-analysis (P = 7.19 × 10−7). Similarly, BIN1 was not statistically significant in our analysis. However, LIMS2, a gene 500 kb upstream of BIN1, was significantly associated (P = 9.43 × 10−12). SNPs in the 3′ untranslated region of LIMS2 have been suggested to associate with cognitive decline52. GWAS index genes for the rest of the loci were all statistically significant in our analysis.
Further, new associations at known risk loci provide novel insights into LOAD etiology. We identified a novel gene, IL10, for LOAD risk (P = 1.77 × 10−7). IL10 is 700 kb upstream of CR1, a strong and consistently replicated locus in LOAD GWAS40,51,53. CR1 was also significant in our analysis (P = 3.71 × 10−7). Although some SNPs near the promoter region of IL10 were moderately associated with LOAD in all three datasets (Supplementary Fig. 9), the IL10–LOAD association was mostly driven by SNPs near CR1 (Supplementary Table 10). An interesting observation is that even when a key SNP is missing—the most significant SNP in IGAP and ADGC (that is, rs2093761:A>G) was not present in GWAX—other predictors (for example, rs6690215:C>T in GWAX) still helped to recover the association signal at the gene level, thus leading to a genome-wide-significant association at IL10. To investigate whether IL10 is simply a companion association signal due to co-regulation with CR1, we performed a cross-tissue conditional analysis using UTMOST with both significant genes CR1 and IL10 included in the model (Methods). Only IL10 remained significant (P = 1.4 × 10−7 for IL10 and P = 0.11 for CR1, Supplementary Table 11) in the conditional analysis. In addition to the strong statistical evidence, the biological function of IL10 also supports its association with LOAD. IL10 is associated with multiple immune diseases54,55,56,57. Moreover, it is known to encode one of the main anti-inflammatory cytokines associated with the occurrence of Alzheimer’s disease and has the therapeutic potential to improve neurodegeneration58,59. Its protein product is also known to physically interact with the Tau protein60.
CLU is another well-replicated risk gene for LOAD. Two independent association peaks at this locus, one at CLU and the other at PTK2B, have been identified in GWAS (Supplementary Fig. 10)40,51. In our analysis, in addition to CLU (P = 1.66 × 10−10), we identified two more significant genes at this locus, ADRA1A (P = 1.29 × 10−9) and EXTL3 (P = 5.08 × 10−12). PTK2B showed marginal association (P = 1.72 × 10−4) with LOAD but did not reach genome-wide significance. Interestingly, EXTL3 expression is predicted by a SNP in the LOAD association peak at CLU, whereas ADRA1A is regulated by SNPs at both CLU and PTK2B (Supplementary Table 12). ADRA1A has been implicated in gene–gene interaction analysis for LOAD61. Its protein product physically interacts with amyloid precursor protein (APP)60, and an α1-adrenoceptor antagonist has been shown to prevent memory deficits in APP23-transgenic mice62. EXTL3 encodes a putative membrane receptor for regenerating islet-derived 1α (Reg-1α), whose overexpression and involvement in the early stages of Alzheimer’s disease has been reported63. Further, the effect of Reg-1α on neurite outgrowth is mediated through EXTL3. Our results provide additional evidence that IL10, ADRA1A, and EXTL3 may be involved in LOAD etiology.
Finally, we identified five novel loci for LOAD, each represented by one significant gene: NICN1 (P = 2.23 × 10−7), RAB43 (P = 1.98 × 10−6), VKORC1 (P = 3.53 × 10−9), HPR (P = 3.02 × 10−7), and PARD6G (P = 3.60 × 10−11). The Rab GTPases are central regulators of intracellular membrane trafficking64. Although RAB43 has not been identified in previous LOAD GWAS, USP6NL, which encodes a GTPase-activating protein for RAB43, has been identified to associate with LOAD in two recent studies45,50. USP6NL also showed suggestive association with LOAD in the discovery stage of our analysis (P = 0.004). However, the associations at RAB43 and USP6NL were not strongly supported by the ADGC or GWAX datasets. Further, the RAB43–LOAD association was driven by SNPs near RPN1, a gene 400 kb downstream of RAB43 (Supplementary Fig. 11 and Supplementary Table 13). This locus is associated with a variety of blood cell traits including monocyte count65,66. VKORC1 is a critical gene in vitamin K metabolism and is the target of warfarin67, a commonly prescribed anticoagulant. The APOE ε4 allele is known to affect the efficacy of warfarin68. HPR has been identified to strongly associate with multiple lipid traits69 and interact with APOE60. NICN1 is known to associate with inflammatory bowel disease70 and cognitive function71. These results provide potential target genes for functional validations in the future. The cross-tissue imputation models of these genes are listed in Supplementary Tables 14–20.
Discussion
Despite the many improvements of UTMOST over existing methods, researchers must be cautious when interpreting findings from UTMOST analyses. First, the gene-level associations identified in UTMOST do not imply causality. It has been recently discussed that correlations among the imputed expression of multiple genes at the same locus may lead to apparent associations at non-causal genes20, which is comparable to the effect of linkage disequilibrium on SNP-level associations in GWAS. Consequently, TWAS-type approaches have limitations in both inferring functional genes and relevant tissues. When eQTL of different genes at the same locus are shared or are in linkage disequilibrium, irrelevant genes may be identified through significant associations. Similarly, for a given gene, if eQTL for the same gene in different tissues are shared or are in linkage disequilibrium, irrelevant tissues may show significant association signals. UTMOST cross-tissue conditional analysis can resolve the issue of gene prioritization to some extent, but fine-mapping of gene-level association remains challenging, especially in regions with extensive linkage disequilibrium. We performed simulations to show that the true associations in the causal tissue were consistently stronger than those in the non-causal tissue in most scenarios, thus indicating that single-tissue association analyses have the potential to enable causal tissue to be inferred (Supplementary Note and Supplementary Fig. 12). However, as the proportion of shared eQTL increased, P values for associations in the non-causal tissue became increasingly significant. Even when two tissues did not share eQTL, associations in the non-causal tissue still frequently passed the significance threshold, probably because of linkage disequilibrium between eQTL. These results are consistent with our experience and discussions in the literature20,72. We also note that these issues may become even more complex when sample sizes and imputation power vary across tissues. Further, we emphasize one of the principles in hypothesis testing—one should not conclude the null hypothesis when an association is not statistically significant. UTMOST is a general framework that involves many analytical steps, and technical issues might mask true gene–trait associations. For example, SPI1 from the CELF1 locus has been causally linked to LOAD risk42. We identified multiple significant associations at this locus, but SPI1 was not a significant gene in our analysis. Possible reasons for this finding include insufficient imputation quality on the basis of the current model, non-availability of causal tissue in the training data, key eQTL missing from the GWAS summary statistics, causal mechanisms (for example, alternative splicing) not well-represented in our analysis, or insufficient sample sizes. In practice, these issues must be carefully investigated before any candidate gene is ruled out.
Overall, UTMOST is a novel, powerful, and flexible framework to perform gene-level association analysis. It integrates biologically informed weights with GWAS summary statistics via modern statistical techniques. If interpreted with caution, its findings may provide insights into disease and trait etiology, motivate downstream functional validation efforts, and eventually benefit the development of novel therapeutics. It is also exciting that statistical and computational methodology in this field evolves rapidly. Several methods of mediation analysis and functional gene fine-mapping in the context of transcriptome-wide association study have been proposed recently73,74. Data-adaptive SNP weights have been shown to effectively improve statistical power at the cost of clear interpretation of associations75. Extension of these methods into multi-tissue analysis is an interesting possible future direction. As high-throughput data continue to be generated for more individuals, cell types, and molecular phenotypes, UTMOST promises to show even better performance and provide greater insights for complex disease genetics in the future.
URLs
UTMOST software, https://github.com/Joker-Jerome/UTMOST/; BLUEPRINT, ftp://ftp.ebi.ac.uk/pub/databases/blueprint/blueprint_Epivar/qtl_as/; STARNET, https://github.com/Wainberg/Vulnerabilities_of_TWAS/; AlzData, http://alzdata.org/index.html; GLGC, http://lipidgenetics.org/; IGAP: http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php/; TWAS summary statistics, ftp://ftp.biostat.wisc.edu/pub/lu_group/Projects/UTMOST/; GEUV, https://www.ebi.ac.uk/arrayexpress/experiments/E-GEUV-1/; GWAX, http://gwas-browser.nygenome.org/downloads/; GTEx, https://www.gtexportal.org; ADGC2 summary statistics, https://www.niagads.org/datasets/ng00076.
Methods
Penalized regression model for cross-tissue expression imputation
Given a gene, we use genotype information to predict its covariate-adjusted expression levels in P tissues. We use SNPs between 1 Mb upstream of the transcription start site and 1 Mb downstream of the transcription end site of the given gene as predictor variables in the model. This is denoted as an N × M matrix X, where N is the total number of individuals, and M denotes the number of SNPs. Throughout the paper, we assume each column of X to be centered but not standardized. Of note, expression data may not be available for all individuals, because only a subset of tissues were collected from each individual. For the ith tissue, we use Ni to denote its sample size. We further use an Ni-dimensional vector Yi to denote the observed expression data in the ith tissue, and we use an Ni × M matrix Xi to denote the genotype information for the subset of individuals. Then, cross-tissue gene expression imputation can be formulated as the following regression problem:
Here, the M × P matrix B summarizes SNPs’ effects on the given gene with its ith column \(B_{ \cdot i}\) denoting the effect sizes of SNPs in the ith tissue and the jth row \(B_{j \cdot }\) denoting the effect sizes of the jth SNP in all P tissues. To effectively select biologically relevant and statistically predictive SNPs, accurately estimate their effects across tissues, and address technical issues including shared samples and incomplete data, we propose the following penalized least-squares estimator for genetic effects matrix B:
Here, ║·║1 and ║·║2 denote the l1 and l2 norms, respectively (that is,║xv×1║1\(= \mathop {\sum}\nolimits_{v = 1}^V {\left| {x_v} \right|}\) and ║xv×1║2\(= \sqrt {\mathop {\sum}\nolimits_{v = 1}^V {x_v^2} }\)). The first term in the loss function is the standard least-squares error. We use the l1 penalty to select predictive variables and impose shrinkage in effect-size estimation. The penalty on each tissue is set adaptively on the basis of the sample sizes, thus reflecting the idea that models for tissues with a larger sample size are more robust to overfitting and therefore are penalized less. To integrate information across multiple tissues, we introduced the third term—a group-lasso penalty on the effect sizes of one SNP29. By imposing this joint penalty across tissues, UTMOST encourages eQTL that are shared across tissues, but it still keeps tissue-specific eQTL with strong effects. Although the penalty on tissue-specific eQTL may cause the model to exclude some true predictors, recent evidence76 has suggested that tissue-specific eQTL have substantially weaker effect sizes and will probably not have major influences on association analysis (Supplementary Note). Tuning parameters λ1 and λ2 control the within-tissue and cross-tissue sparsity, respectively. They are selected through cross-validation. Details of the optimization can be found in the Supplementary Note.
Model training and evaluation
We trained our cross-tissue gene expression imputation model by using genotype and normalized gene expression data from 44 tissues in the GTEx project (version V6p, dbGaP accession code phs000424.v6.p1)3. The sample sizes for different tissues ranged from 70 (uterus) to 361 (skeletal muscle). SNPs with ambiguous alleles or minor allele frequency (MAF) <0.01 were removed. Normalized gene expression was further adjusted to remove potential confounding effects from sex, sequencing platform, the top three principal components of genotype data, and the top probabilistic estimation of expression residuals (PEER) factors77. As previously recommended17, we included 15 PEER factors for tissues with N <150, 30 factors for tissues with 150 ≤ N < 250, and 35 factors for tissues with N ≥250. All covariates were downloaded from the GTEx portal website (see URLs). We applied a fivefold cross-validation for model tuning and evaluation. Specifically, we randomly divided individuals into five groups of equal size. Each time, we used three groups as the training set, one as the intermediate set for selecting tuning parameters, and the last one as the testing set for performance evaluation. The squared correlation between predicted and observed expression (that is, R2) was used to quantify imputation accuracy. For each model, we selected gene–tissue pairs with FDR <0.05 for downstream testing. External validation of imputation accuracy was performed by using whole-blood expression data from 421 samples in the 1,000 Genomes Project (GEUVADIS consortium)32 and the CommonMind consortium33, which collected expression in across multiple regions from >1,000 postmortem brain samples (mainly corresponding to Brain_Frontal_Cortex_BA9 in GTEx) from donors with schizophrenia or bipolar disorder, and individuals with no neuropsychiatric disorders. For CommonMind data, we focused our analysis on 147 controls with no neuropsychiatric disorders. Average improvements in R2 in both external validation datasets are shown in Supplementary Fig. 4. Although not statistically significant, owing to the limited sample size, the accuracy of the cross-tissue method was consistently higher than that of the single-tissue approach in different quantiles. Furthermore, comparing the tissue–tissue similarity on the basis of the observed and imputed gene expressions indicated that cross-tissue imputation removed stochastic noises in the expression data without losing tissue-specific correlational patterns (Supplementary Note and Supplementary Figs. 5 and 6).
Gene-level association test
We combined GWAS summary statistics with SNP effects estimated in the cross-tissue imputation model (that is, \(\hat B\)) to quantify gene–trait associations in each tissue. For a given gene, we modeled its imputed expression in the ith tissue (that is, \(E_i = X_i\hat B_{ \cdot i}\)) and the phenotype T by using a linear model:
Then, the association statistic for effect size in the ith tissue (that is, γi) on the trait of interest is:
where \(\hat \gamma _i\) denotes the point estimate for effect size, and (s.e.m.\(\hat \gamma _i\)) denotes its standard error of the mean. From the linear model, we have:
where Γi is an M × M diagonal matrix with the jth term equal to \(\frac{{\sigma _j}}{{\eta _i}}\), where σj is the standard deviation of the jth SNP, and ηi is the standard deviation of imputed gene expression in the ith tissue. These parameters could be estimated by using a reference panel. \(\tilde \beta\) denotes the SNP-level effect-size estimates acquired from GWAS summary statistics. Regarding the standard error of the mean of \(\hat \gamma _i\), we have:
Here, σY denotes the standard deviation of phenotype T, and NGWAS is the sample size in GWAS. The approximation \(\mathrm{var}(\delta _i) \approx \sigma _Y^2\) is based on the empirical observation that each gene explains only a very small proportion of phenotypic variability78. The same argument can be extended to association statistics at the SNP level. For the jth SNP in the model, we have
Therefore, SNP-level z scores can be denoted as:
In matrix form, this is:
Combining the derivations above, we can denote the gene-level z score as:
Under the null hypothesis (that is no SNP-trait association), \(\tilde Z\) follows a multivariate normal distribution \(\tilde Z\approx N(0,D)\), where D is the linkage-disequilibrium matrix for SNPs and could be estimated by using an external reference panel. Denoting the cross-tissue gene–trait z scores as Z = (Z1, Z2, …, ZP)T, the covariance matrix of Z could be calculated as:
where \({\mathrm{\Lambda }} = (\hat B_{ \cdot 1}{\mathrm{\Gamma }}_1,\,\hat B_{ \cdot 2}{\mathrm{\Gamma }}_2,\, \ldots ,\,\hat B_{ \cdot P}{\mathrm{\Gamma }}_P)\).
To combine gene–trait associations across multiple tissues, we applied the GBJ test with single-tissue association statistics Z and their covariance matrix Σ as inputs. This approach provides powerful inference results while explicitly taking the correlation among single-tissue test statistics into account even under a sparse alternative (that is, biologically meaningful associations are present in only a small number tissues)30. The GBJ test statistic can be calculated as:
where |Z|(i) denotes the ith order statistic of the absolute value of gene–trait z scores in an increasing order; \(S\left( t \right) = \mathop {\sum}\nolimits_{i = 1}^P 1 \left( {\left| {Z_i} \right| \ge t} \right)\) denotes the number of gene–trait z scores with absolute value greater than a threshold t; \(\hat \mu _i\) denotes the corresponding value of E(Z) that maximizes the probability of event S(|Z|(P−i + 1)) = i; and \({\bar{\mathrm \Phi }}\left( t \right) = 1 - {\mathrm{\Phi }}\left( t \right)\) is the survival function of the standard normal distribution. The GBJ test statistic can be interpreted as the maximum of a series of one-sided likelihood-ratio test statistics on the mean of S(t), where the denominator denotes the maximum likelihood when no gene–trait association exists in any tissue (all z scores have zero mean), and the numerator denotes the unconstrained maximum likelihood. Of note, calculating the exact distribution of S(t) is difficult when z scores are correlated. As previously suggested, we calculate G by approximating the distribution of S(t) with an extended beta-binomial (EBB) distribution. As a maximum-based global statistic, the P value of the GBJ test could be written as:
where 0 ≤ b1 ≤ b2 ≤ … ≤ bP are ‘boundary points’ derived from inversion of the test statistic, which depends on G, P, and Σ. MVN denotes multivariatte normal distribution. The last quantity in the equation can be calculated recursively with the EBB approximation30.
P-value cutoffs for gene-level association tests were determined by Bonferroni correction. For each method, we used 0.05 divided by the total number of genes tested across 44 tissues (that is, 5.76 × 10−7 for TWAS, 2.44 × 10−7 for PrediXcan, and 1.28 × 10−7 for UTMOST, respectively) as the significance threshold. As more genes can be accurately imputed (R2 significantly larger than zero with FDR <0.05) in our cross-tissue imputation, the significance cutoff was the most stringent in UTMOST.
Cross-tissue conditional analysis
Genes that are physically close to the true risk gene may be identified in marginal association analyses, owing to co-regulation of multiple genes by the same eQTL and linkage disequilibrium between eQTL of different genes. To prioritize gene-level associations at the same locus, we expand UTMOST to perform cross-tissue conditional analysis. There are two major steps in this framework.
First, at any pre-defined locus, we can derive the formula of conditional analysis on the basis of marginal associations. T denotes the trait of interest. The goal is to perform a multiple regression analysis using K imputed gene expressions in the ith tissue (that is, Ei1,…, EiK) as predictor variables:
Here, we use \(E_i^ \ast = (E_{i1}, \ldots ,\,E_{iK})\) to denote an N × K matrix for K imputed gene expressions in the ith tissue. Regression coefficients \(\gamma _i^ \ast = (\gamma _{i1}, \ldots ,\gamma _{iK})^{\mathrm{T}}\) are the parameters of interest. To simplify the algebra, we also assume that trait T and all SNPs in the genotype matrix X are centered so there is no intercept term in the model, but the conclusions apply to the general setting. Similarly to univariate analysis, gene expression levels Ei1,…,EiK are imputed from genetic data via linear prediction models:
where \(B_i^ \ast\) are imputation weights assigned to SNPs. The kth column of \(B_i^ \ast\) denotes the imputation model for gene expression Eik. Then, the ordinary least-squares estimator \(\hat \gamma ^ \ast\) and its variance–covariance matrix can be denoted as follows:
The approximation is based on the assumption that imputed gene expression levels Ei1, …, EiK collectively explain little variance in T, which is reasonable in complex gene expression genetics if K is not large. We further denote:
All elements in matrix Ui can be approximated by using a reference panel \(\tilde X\). Therefore, the z score for γik(1 ≤ k ≤ K) is:
where Ik is the K × 1 vector with the kth element being 1 and all other elements equal to 0, Θ is an M × M diagonal matrix with the jth diagonal element being \(\sqrt {\mathrm{var}\left( {X_j} \right)}\), and, similarly to the notation in univariate analysis, \(\tilde Z\) is the vector of SNP-level z scores from the GWAS of trait T. Importantly, we note that given imputation models for K gene expressions (that is, \(B_i^ \ast\)), GWAS summary statistics for trait T (that is, \(\tilde Z\)), and an external genetic dataset to estimate Ui and Θ, conditional analysis can be performed without individual-level genotype and phenotype data.
In the second step, we combine the conditional analysis association statistics across different tissues by using the GBJ test. Of note, this is different from the final stage of UTMOST, which combines the marginal gene–trait-tissue associations. Through these two steps, linkage disequilibrium between eQTL and co-regulation across tissues has been taken into account in the test. Specifically, under the null hypothesis (that is, no SNP-trait association), \(\tilde Z\) follows a multivariate normal distribution \(\tilde Z\sim N(0,D)\), where D is the linkage-disequilibrium matrix for SNPs and could be estimated by using an external reference panel. By denoting the cross-tissue gene–trait z scores for gene k as Zk = (Z1k,Z2k,…,ZPk)T, the covariance matrix of Zk could be calculated as:
where
Simulation settings
Genotype data from 12,637 individuals in the GERA dataset (dbGaP accession phs000674), including 7,432 type 2 diabetes cases (phenotypic information not used) and 5,205 healthy controls, were used in the simulation studies. We removed SNPs with missing rate above 0.01 and individuals with genetic relatedness coefficients above 0.05. The genotype data were imputed to the 1,000 Genomes Project Phase 1v3 European samples by using the Michigan Imputation Server79. After imputation, we further removed SNPs with MAF <0.05. After quality control, 5,932,546 SNPs remained in the dataset.
We performed two different simulation studies to evaluate the type I error rate of our cross-tissue association test. First, we directly simulated quantitative traits from a standard normal distribution independent from the genotype data, and we then performed single-tissue association tests for 44 tissues in GTEx and the GBJ cross-tissue association test for all genes, by using the simulated data. In the second setting, we simulated genetically regulated expression components and then simulated the GWAS trait on the basis of gene expression values. For each gene, we simulated its expression in three tissues, namely skeletal muscle (N = 361), skin from sun-exposed lower leg (N = 302) and whole blood (N = 338). Within the ith tissue, the cis component of gene expression was generated as \(E_i = X_i\hat B_{ \cdot i}\). We used real effect sizes \(\hat B_{ \cdot i}\) estimated in our joint imputation model so that the genetic architecture of gene expression was preserved in the simulations. Next, the quantitative trait value was simulated as Y = w1E1 + w2E2 + w3E3 + ε, where wi is the effect of gene expression on the trait in the ith tissue. To evaluate type I error, we set w1 = w2 = w3 = 0, that is, none of the three tissues are relevant to the trait.
To simulate data under the alternative hypothesis, we generated diverse disease architectures by considering different numbers of causal tissues (that is, 1, 2, or 3 causal tissues) and two heritability settings (that is, 0.01 and 0.001). Specifically, we fixed the total variance explained by E1, E2, and E3 and varied wi to simulate different levels of tissue specificity of the trait. We generated traits by using the following three settings:
-
Setting 1. w1 = 1,w2 = w3 = 0. Only the first tissue contributes to the disease; the other two tissues are not relevant.
-
Setting 2. \(w_1 = w_2 = \frac{1}{2},\,w_3 = 0\). Both the first and the second tissue contribute equally to disease; the third tissue is irrelevant to the disease.
-
Setting 3. \(w_1 = w_2 = w_3 = \frac{1}{3}\). All three tissues contribute equally to the disease.
Single-tissue and cross-tissue gene–trait associations were then estimated by using the UTMOST framework. We repeated the entire procedure on 200 randomly selected genes. For each gene, we further replicated five times. Statistical power was calculated as the proportion of test P values reaching the significance threshold, that is, 0.05/15,000 for both single-tissue and cross-tissue tests and 0.05/45,000 for single-tissue tests while accounting for the number of tissues.
GWAS data analysis
We applied UTMOST to GWAS summary statistics for 50 complex diseases and traits. Details of these 50 studies are summarized in Supplementary Table 4. GWAS summary statistics for LDL-C were downloaded from the Global Lipids Genetics Consortium website (see URLs). Summary statistics from the IGAP stage I analysis were downloaded from the IGAP website (see URLs). The GWAX result for LOAD was downloaded from the New York Genome Center website (see URLs). ADGC phase 2 summary statistics were generated by first analyzing individual datasets by using logistic regression adjusting for age, sex, and the first three principal components in the program SNPTest v.2 (ref. 80). Meta-analysis of the individual dataset results was then performed by using the inverse-variance weighted approach in METAL81.
To identify trait-related tissue, we first used GenoSkyline-Plus, an unsupervised learning framework trained on various epigenetic marks from the Roadmap Epigenomics Project82, to quantify tissue-specific functionality in the human genome83. We then estimated the enrichment for trait heritability in each tissue’s predicted functional genome by using linkage-disequilibrium-score regression34. More specifically, annotation-stratified linkage-disequilibrium scores were estimated by using the 1,000 Genomes samples of European ancestry and a 1-cM window. GenoSkyline-Plus annotations for 27 tissues that can be matched between Roadmap and GTEx were included in the linkage-disequilibrium-score regression model together with 53 baseline annotations, as previously suggested34. For each tissue-specific annotation, partitioned heritability was estimated, and enrichment was calculated as the ratio of the proportion of explained heritability and the proportion of SNPs in each annotated category. Tissue-trait relevance was then ranked according to enrichment P values. We used the term ‘most enriched tissues’ to denote the tissues that were most significantly enriched for heritability of each trait. Finucane et al.84 have also applied LD-score regression (LDSC) with tissue-specific annotations based on GTEx data to infer trait-related tissues. Because UTMOST was based on GTEx data, we used independent data from the Roadmap project to infer trait-relevant tissues for the purpose of fair comparison.
In the UTMOST analytical framework, multiple parameters must be estimated by using an external reference panel (for example, linkage disequilibrium). We used samples with European ancestry from the 1,000 Genomes Project for this estimation85. When performing cross-tissue association tests, we combined single-tissue statistics from tissues that passed FDR <0.05 criteria to reduce noise in the analysis. Genome-wide significance was defined as 3.3 × 10−6 (that is, Bonferroni correction based on 15,120 genes that passed the quality control steps). For heritability enrichment analysis, we applied LDSC to 27 GenoSkyline-Plus tissue-specific annotations with matched tissue types in GTEx (Supplementary Table 21). The 53 LDSC baseline annotations were also included in the model, as previously recommended34. The most and least relevant tissues were selected according to the enrichment-test P values. Gene ontology enrichment analysis was performed by using DAVID86. Protein–protein-interaction information was acquired from the AlzData website (see URLs)60. Locus plots for SNP-level GWAS associations were generated by using LocusZoom87. Manhattan plots were generated by using the qqman package in R88.
Additional QTL data
The imputation model for liver tissue in the STARNET study (N = 522) was downloaded from the Github page (see URLs). Predictor effects were trained by using an elastic net model with variants within a 500-kb range of the transcription start site. Details of the quality control procedure have been previously reported20. We also collected additional eQTL and splicing QTL (sQTL) data for three immune cell types (CD14+ monocytes, CD16+ neutrophils, and naive CD4+ T cells; 169–194 samples per tissue) from the BLUEPRINT consortium (see URLs). eQTLs with FDR <0.01 and sQTLs with FDR <0.05 were used in the gene-level association analysis for LOAD.
We also downloaded monocyte eQTL summary statistics from the Immune Variation Project89 for comparison with BLUEPRINT results in LOAD. We first compared the monocyte eQTL identified in BLUEPRINT with what was identified in this dataset (denote as ImmVar). Only a very low fraction (3.5%) of the eQTLs was replicated in ImmVar. We further performed single-tissue analysis on LOAD with weights constructed from ImmVar and compared the identified associations with those identified by using BLUEPRINT data (Supplementary Tables 22 and 23). Significant genes did not match between the two analyses, probably because of the small overlap of eQTLs between the two datasets. However, UTMOST uses the GBJ statistic to combine associations across datasets and therefore has the flexibility to incorporate single-tissue associations based on external eQTL studies. As we demonstrated in the case study of LDL-C at the SORT1 locus, incorporating STARNET liver eQTL significantly increased the statistical power although liver was an available tissue in GTEx. As sample sizes and tissue types in QTL studies continue to grow, UTMOST will be able to incorporate additional data sources and provide better results.
Statistical tests
We tested the difference in R2 across genes with one-sided Kolmogorov–Smirnov test, which calculates the largest distance between the empirical cumulative distribution functions and uses it to test whether two distributions are identical (Supplementary Fig. 3 and 4). Furthermore, we used a paired Wilcoxon rank test to compare the number of genes identified in different tissues between different methods, which is a non-parametric test used to compare two matched samples to access whether their population means differ (Fig. 4 and Supplementary Fig. 7).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data used in the manuscript are publicly available (see URLs). GTEx and GERA data can be accessed by application to dbGaP. CommonMind data are available through formal application to NIMH. ADGC phase 2 summary statistics used for validation are available through the NIAGADS portal under accession number NG00076.
References
Visscher, P. M. et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017).
Boyle, E. A., Li, Y. I. & Pritchard, J. K. An Expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 (2017).
Ardlie, K. G. et al. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Aguet, F. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Yang, F. et al. Identifying cis-mediators for trans-eQTLs across many human tissues using genomic mediation analysis. Genome Res. 27, 1859–1871 (2017).
Saha, A. et al. Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res. 27, 1843–1858 (2017).
Mohammadi, P., Castel, S. E., Brown, A. A. & Lappalainen, T. Quantifying the regulatory effect size of cis-acting genetic variation using allelic fold change. Genome Res. 27, 1872–1884 (2017).
Nicolae, D. L. et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS. Genet. 6, e1000888 (2010).
Hou, L., Chen, M., Zhang, C. K., Cho, J. & Zhao, H. Guilt by rewiring: gene prioritization through network rewiring in genome wide association studies. Hum. Mol. Genet. 23, 2780–2790 (2013).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Hormozdiari, F. et al. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 99, 1245–1260 (2016).
Zhao, S. D., Cai, T. T., Cappola, T. P., Margulies, K. B. & Li, H. Sparse simultaneous signal detection for identifying genetically controlled disease genes. J. Am. Stat. Assoc. 112, 1032–1046 (2016).
Gamazon, E. R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Mancuso, N. et al. Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am. J. Hum. Genet. 100, 473–487 (2017).
Barbeira, A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Hoffman, J. D. et al. Cis-eQTL-based trans-ethnic meta-analysis reveals novel genes associated with breast cancer risk. PLoS Genet. 13, e1006690 (2017).
Liu, X. et al. Functional architectures of local and distal regulation of gene expression in multiple human tissues. Am. J. Hum. Genet. 100, 605–616 (2017).
Wainberg, M. et al. Vulnerabilities of transcriptome-wide association studies. Preprint at https://www.biorxiv.org/content/10.1101/206961v5 (2017).
Li, C., Yang, C., Gelernter, J. & Zhao, H. Improving genetic risk prediction by leveraging pleiotropy. Hum. Genet. 133, 639–650 (2014).
Maier, R. et al. Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder, and major depressive disorder. Am. J. Hum. Genet. 96, 283–294 (2015).
Hu, Y. et al. Joint modeling of genetically correlated diseases and functional annotations increases accuracy of polygenic risk prediction. PLoS Genet. 13, e1006836 (2017).
Flutre, T., Wen, X., Pritchard, J. & Stephens, M. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet. 9, e1003486 (2013).
Sul, J. H., Han, B., Ye, C., Choi, T. & Eskin, E. Effectively identifying eQTLs from multiple tissues by combining mixed model and meta-analytic approaches. PLoS Genet. 9, e1003491 (2013).
Duong, D. et al. Applying meta-analysis to genotype-tissue expression data from multiple tissues to identify eQTLs and increase the number of eGenes. Bioinformatics 33, i67–i74 (2017).
Li, G., Jima, D. D., Wright, F. A. & Nobel, A. B. HT-eQTL: integrative eQTL analysis in a large number of human tissues. BMC Bioinformatics 19, 95 (2018).
Hore, V. et al. Tensor decomposition for multiple-tissue gene expression experiments. Nat. Genet. 48, 1094–1100 (2016).
Yuan, M. & Lin, Y. Model selection and estimation in regression with grouped variables. J. Royal Stat. Soc. B 68, 49–67 (2006).
Sun, R. & Lin, X. Set-based tests for genetic association using the generalized Berk–Jones statistic. Preprint at https://arxiv.org/pdf/1710.02469 (2017).
Zhou, X., Carbonetto, P. & Stephens, M. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 9, e1003264 (2013).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506 (2013).
Fromer, M. et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 19, 1442 (2016).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228 (2015).
Lu, Q. et al. Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease. PLoS Genet. 13, e1006933 (2017).
Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Franzén, O. et al. Cardiometabolic risk loci share downstream cis-and trans-gene regulation across tissues and diseases. Science 353, 827–830 (2016).
Musunuru, K. et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466, 714–719 (2010).
Strong, A. et al. Hepatic sortilin regulates both apolipoprotein B secretion and LDL catabolism. J. Clin. Invest. 122, 2807 (2012).
Lambert, J.-C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458 (2013).
Gagliano, S. A. et al. Genomics implicates adaptive and innate immunity in Alzheimer’s and Parkinson’s diseases. Ann. Clin. Transl. Neurol. 3, 924–933 (2016).
Huang, K. L. et al. A common haplotype lowers PU. 1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 20, 1052 (2017).
Raj, T. et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet. 50, 1584 (2018).
Chen, L. et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167, 1398–1414. e24 (2016).
Liu, J. Z., Erlich, Y. & Pickrell, J. K. Case-control association mapping by proxy using family history of disease. Nat. Genet. 49, 325–331 (2017).
Hollingworth, P. et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 43, 429–435 (2011).
Harold, D. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 41, 1088–1093 (2009).
Naj, A. C. et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 43, 436–441 (2011).
Seshadri, S. et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303, 1832–1840 (2010).
Jun, G. R. et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement. 13, 727–738 (2017).
Lambert, J. C. et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 41, 1094–1099 (2009).
Sherva, R. et al. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimers Dement. 10, 45–52 (2014).
Crehan, H. et al. Complement receptor 1 (CR1) and Alzheimer’s disease. Immunobiology 217, 244–250 (2012).
Liu, J. Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015).
Remmers, E. F. et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet’s disease. Nat. Genet. 42, 698–702 (2010).
Plagnol, V. et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 7, e1002216 (2011).
Bentham, J. et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 47, 1457–1464 (2015).
Kiyota, T. et al. AAV serotype 2/1-mediated gene delivery of anti-inflammatory interleukin-10 enhances neurogenesis and cognitive function in APP + PS1 mice. Gene Ther. 19, 724–733 (2012).
Chakrabarty, P. et al. IL-10 alters immunoproteostasis in APP mice, increasing plaque burden and worsening cognitive behavior. Neuron 85, 519–533 (2015).
Xu, M. et al. A systematic integrated analysis of brain expression profiles reveals YAP1 and other prioritized hub genes as important upstream regulators in Alzheimer’s disease. Alzheimers Dement. 14, 215–229 (2017).
Hohman, T. J. et al. Discovery of gene–gene interactions across multiple independent data sets of late onset Alzheimer disease from the Alzheimer Disease Genetics Consortium. Neurobiol. Aging 38, 141–150 (2016).
Katsouri, L. et al. Prazosin, an α 1-adrenoceptor antagonist, prevents memory deterioration in the APP23 transgenic mouse model of Alzheimer’s disease. Neurobiol. Aging 34, 1105–1115 (2013).
Duplan, L. et al. Lithostathine and pancreatitis-associated protein are involved in the very early stages of Alzheimer’s disease. Neurobiol. Aging 22, 79–88 (2001).
Stenmark, H. & Olkkonen, V. M. The rab gtpase family. Genome. Biol. 2, reviews3007 (2001).
Lin, B. D. et al. Heritability and GWAS studies for monocyte–lymphocyte ratio. Twin Res Hum. Genet. 20, 97–107 (2017).
Astle, W. J. et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167, 1415–1429.e19 (2016).
Li, T. et al. Identification of the gene for vitamin K epoxide reductase. Nature 427, 541–544 (2004).
Kohnke, H., Sörlin, K., Granath, G. & Wadelius, M. Warfarin dose related to apolipoprotein E (APOE) genotype. Eur. J. Clin. Pharmacol. 61, 381–388 (2005).
Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
de Lange, K. M. et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49, 256–261 (2017).
Davies, G. et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N = 53 949). Mol. Psychiatry 20, 183 (2015).
Torres, J. M. et al. Integrative cross tissue analysis of gene expression identifies novel type 2 diabetes genes. Preprint at https://www.biorxiv.org/content/10.1101/108134v2 (2017).
Park, Y. et al. Causal gene inference by multivariate mediation analysis in Alzheimer’s disease. Preprint at https://www.biorxiv.org/content/10.1101/219428v3 (2017).
Mancuso, N. et al. Probabilistic fine-mapping of transcriptome-wide association studies. Preprint at https://www.biorxiv.org/content/10.1101/236869v2 (2017).
Xu, Z., Wu, C., Wei, P. & Pan, W. A powerful framework for integrating eQTL and GWAS summary data. Genetics 207, 893–902 (2017).
GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Stegle, O., Parts, L., Durbin, R. & Winn, J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput. Biol. 6, e1000770 (2010).
O’Connor, L. J. et al. Estimating the proportion of disease heritability mediated by gene expression levels. Preprint at https://www.biorxiv.org/content/10.1101/118018v1 (2017).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284 (2016).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317 (2015).
Lu, Q. et al. Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease. PLoS Genet. 13, e1006933 (2017).
Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621 (2018).
Abecasis, G. R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Turner, S. D. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. Preprint at https://www.biorxiv.org/content/10.1101/005165v1 (2014).
Raj, T. et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523 (2014).
Acknowledgements
This study was supported in part by NIH grants R01 GM59507 and 3P30AG021342-16S2 (Y.H., M.L., Q.L., and H.Z.), CTSA UL1TR000427 (Q.L.), R01 AG042437 and U01 AG006781 (P.K.C. and S. Mukherjee); the Yale World Scholars Program sponsored by the China Scholarship Council (J.W., and Z.Y.); Neil Shen’s SJTU Medical Research Fund, the SJTU-Yale Collaborative Research Seed Fund; and NSFC 31728012 (J.G., H.L., and H.Z.), and the National Key R&D Program of China 2018YFC0910500 (J.G., and H.L). We thank C. Brown for assistance in matching GTEx tissues to Roadmap cell types. This study makes use of summary statistics from many GWAS consortia. We thank the investigators in these GWAS consortia for generously sharing their data. We thank the IGAP for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in the analysis or writing of this report. IGAP was made possible by the generous participation of the subjects and their families. The i-Select chips were funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (Laboratory of Excellence Program Investment for the Future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2, and the Lille University Hospital. The Genetic and Environmental Risk in AD consortium (GERAD) was supported by the Medical Research Council (grant no. 503480), Alzheimer’s Research UK (grant no. 503176), the Wellcome Trust (grant no. 082604/2/07/Z), and the German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant nos. 01GI0102, 01GI0711, and 01GI0420. The Cohorts for Heart and Aging Research in Genomic Epidemiology consortium (CHARGE) was partly supported by NIH/NIA grant no. R01 AG033193, NIA grant no. AG081220, AGES contract N01–AG–12100, NHLBI grant no. R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by NIH/NIA grants nos. U01 AG032984, U24 AG021886, and U01 AG016976, and the Alzheimer’s Association grant no. ADGC–10–196728. We thank the contributors who collected the samples used in this study, as well as the patients and their families, whose help and participation made this work possible; data for this study were prepared, archived, and distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (U24-AG041689-01). We are also grateful for all the consortia and investigators that provided publicly accessible GWAS summary statistics.
Author information
Authors and Affiliations
Consortia
Contributions
Y.H., M.L., Q.L., H.L., and H.Z. conceived the study and developed the statistical model. Y.H., M.L., Q.L., H.W., J.W., S.M.Z., B.L., Y.S., S. Muchnik, and J.G. performed the statistical analyses. S.M.Z. and P.N. assisted in LDL analysis. Y.H., M.L., Z.Y., and Q.L. implemented the software. B.W.K. prepared ADGC summary statistics. A.N., A.K., and Y.Z. assisted in data preparation. S. Mukherjee and P.K.C. assisted in Alzheimer’s disease data application, curation, and interpretation. Y.H., M.L., Q.L., H.L., and H.Z. wrote the manuscript. H.Z. advised on statistical and genetic issues. All authors contributed to manuscript editing and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Tables 1–13, 21–23 and 26–29, and Supplementary Figures 1–13
Supplementary Tables
Supplementary Tables 14–20, 24 and 25
Rights and permissions
About this article
Cite this article
Hu, Y., Li, M., Lu, Q. et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat Genet 51, 568–576 (2019). https://doi.org/10.1038/s41588-019-0345-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-019-0345-7
This article is cited by
-
Integrating human plasma proteomes with genome-wide association data implicates novel proteins and drug targets for rheumatoid arthritis
Clinical Proteomics (2026)
-
Genetic insights and mechanistic parallels in gestational diabetes mellitus and type 2 diabetes
Nature Communications (2026)
-
Unveiling novel susceptibility genes for kidney stone disease by a cross-tissue transcriptome-wide association study
World Journal of Urology (2026)
-
Transcriptome remodeling of mouse hearts during postnatal cardiac maturation and under proteotoxic stress
Molecular Biology Reports (2026)
-
A cross-tissue transcriptome-wide association study identifies novel candidate genes associated with brain glymphatic system function
Molecular Brain (2025)