Abstract
Drought poses a severe threat to the stability of crop yields. It is crucial to identify genetic resources and decipher the molecular mechanisms underlying drought resistance in crops. Here we generated high-quality genome assemblies of 25 maize germplasms exhibiting substantial variation in drought resistance. Combined with 31 additional maize genome sequences, a comprehensive pangenome analysis was performed. Rare allelic variations and extensive regulatory diversity were revealed in abscisic acid-related or drought-related genes, which may contribute to the diversity in drought resistance among germplasms. Furthermore, we identified three genes, ZmUGE2, ZmSIL2 and ZmASI3, that enhance maize drought resistance by strengthening mechanical support of the cell wall, regulating stress-responsive gene expression and coordinating male and female inflorescence development, respectively. Thus, this study provides valuable insight into the genetic control of drought resistance in maize at different growing stages. The expanded maize pangenome information serves as a valuable resource for maize genomic research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All raw sequencing data referenced in this study are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive with BioProject accession number PRJNA1103102, including the ONT, DNA and/or RNA Illumina sequencing, as well as RNA-seq and ChIP–seq data for transgenic plants. Genome sequences and annotations for 25 newly assembled maize germplasms and SV information are available via Zenodo at https://doi.org/10.5281/zenodo.16576184 (ref. 99). GWAS results related to ZmUGE2, ZmSIL2 and ZmASI3 are available via Zenodo at https://doi.org/10.5281/zenodo.17138577 (ref. 100). Long-read sequencing data of the previously published genomes were obtained from the NCBI database, which includes NAM founders16 (accession number PRJEB31061), Mo1727 (accession number PRJNA358298), SK31 (accession number PRJNA531547), CIMBL5525 (accession number PRJNA765111), K0326Y32 (accession number PRJNA539996) and A18833 (accession number PRJNA635654). Other public maize, teosinte and T. dactyloides genome sequences used in this work are available at https://download.maizegdb.org/. Source data are provided with this paper.
Code availability
All customized scripts used in this study are available via GitHub at https://github.com/YangBioinformatics/Maize-Pan-genome and via Zenodo at https://doi.org/10.5281/zenodo.16631382 (ref. 101).
References
The Impact of Disasters and Crises on Agriculture and Food Security (FAO, 2021).
Erenstein, O., Chamberlin, J. & Sonder, K. Estimating the global number and distribution of maize and wheat farms. Glob. Food Secur. 30, 100558 (2021).
Harrison, M. T., Tardieu, F., Dong, Z., Messina, C. D. & Hammer, G. L. Characterizing drought stress and trait influence on maize yield under current and future conditions. Glob. Chang. Biol. 20, 867–878 (2014).
Lobell, D. B., Deines, J. M. & Tommaso, S. D. Changes in the drought sensitivity of US maize yields. Nat. Food 1, 729–735 (2020).
Lobell, D. B. et al. Greater sensitivity to drought accompanies maize yield increase in the U.S. Midwest. Science 344, 516–519 (2014).
Chen, L. et al. Genome sequencing reveals evidence of adaptive variation in the genus Zea. Nat. Genet. 54, 1736–1745 (2022).
Wang, X. et al. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 48, 1233–1241 (2016).
Bolaños, J. & Edmeades, G. O. The importance of the anthesis-silking interval in breeding for drought tolerance in tropical maize. Field Crops Res. 48, 65–80 (1996).
Liu, B. et al. Manipulating ZmEXPA4 expression ameliorates the drought-induced prolonged anthesis and silking interval in maize. Plant Cell 33, 2058–2071 (2021).
Danilevskaya, O. N. et al. Developmental and transcriptional responses of maize to drought stress under field conditions. Plant Direct 3, e00129 (2019).
Fuad-Hassan, A., Tardieu, F. & Turc, O. Drought-induced changes in anthesis-silking interval are related to silk expansion: a spatio-temporal growth analysis in maize plants subjected to soil water deficit. Plant Cell Environ. 31, 1349–1360 (2008).
Liu, S. et al. Mapping regulatory variants controlling gene expression in drought response and tolerance in maize. Genome Biol. 21, 163 (2020).
Mao, H. et al. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 6, 8326 (2015).
Ricci, W. A. et al. Widespread long-range cis-regulatory elements in the maize genome. Nat. Plants 5, 1237–1249 (2019).
Jiao, Y. et al. Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017).
Hufford, M. B. et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science 373, 655–662 (2021).
Alonge, M. et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182, 145–161 (2020).
Liu, Y. C. et al. Pan-genome of wild and cultivated soybeans. Cell 182, 162–176 (2020).
Qin, P. et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184, 3542–3558 (2021).
Wang, B. et al. De novo genome assembly and analyses of 12 founder inbred lines provide insights into maize heterosis. Nat. Genet. 55, 312–323 (2023).
Jayakodi, M. et al. Structural variation in the pangenome of wild and domesticated barley. Nature 636, 654–662 (2024).
Jiao, C. et al. Pan-genome bridges wheat structural variations with habitat and breeding. Nature 637, 384–393 (2024).
Gui, S. et al. A pan-Zea genome map for enhancing maize improvement. Genome Biol. 23, 178 (2022).
Zhou, Y. et al. Graph pangenome captures missing heritability and empowers tomato breeding. Nature 606, 527–534 (2022).
Tian, T. et al. Genome assembly and genetic dissection of a prominent drought-resistant maize germplasm. Nat. Genet. 55, 496–506 (2023).
Yang, X. et al. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol. Breed. 28, 511–526 (2010).
Sun, S. et al. Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat. Genet. 50, 1289–1295 (2018).
Chen, J. et al. A complete telomere-to-telomere assembly of the maize genome. Nat. Genet. 55, 1221–1231 (2023).
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Ou, S., Chen, J. & Jiang, N. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res. 46, e126 (2018).
Yang, N. et al. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat. Genet. 51, 1052–1059 (2019).
Li, C. et al. Long-read sequencing reveals genomic structural variations that underlie creation of quality protein maize. Nat. Commun. 11, 17 (2020).
Lin, G. et al. Chromosome-level genome assembly of a regenerable maize inbred line A188. Genome Biol. 22, 175 (2021).
Huang, Y. et al. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 287, 110188 (2019).
Uga, Y. et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102 (2013).
Zhang, F. et al. Genomic basis underlying the metabolome-mediated drought adaptation of maize. Genome Biol. 22, 260 (2021).
Wu, X. et al. Using high-throughput multiple optical phenotyping to decipher the genetic architecture of maize drought tolerance. Genome Biol. 22, 185 (2021).
Zhang, Y. et al. BRITTLE PLANT1 is required for normal cell wall composition and mechanical strength in rice. J. Integr. Plant Biol. 63, 865–877 (2021).
Liu, S. et al. The rice BZ1 locus is required for glycosylation of arabinogalactan proteins and galactolipid and plays a role in both mechanical strength and leaf color. Rice 13, 41 (2020).
Rosti, J. et al. UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell 19, 1565–1579 (2007).
Kaplan-Levy, R. N., Brewer, P. B., Quon, T. & Smyth, D. R. The trihelix family of transcription factors—light, stress and development. Trends Plant Sci. 17, 163–171 (2012).
Yang, N. et al. Two teosintes made modern maize. Science 382, eadg8940 (2023).
Sakamoto, H. et al. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136, 2734–2746 (2004).
Liu, S. et al. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 9, e1003790 (2013).
Wu, Q. et al. Transcription factor ZmEREB97 regulates nitrate uptake in maize (Zea mays) roots. Plant Physiol. 196, 535–550 (2024).
Huh, S. U. New function of hypoxia-responsive unknown protein in enhanced resistance to biotic stress. Plant Signal Behav. 16, 1868131 (2021).
Knizewski, L., Ginalski, K. & Jerzmanowski, A. Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 13, 557–565 (2008).
Deng, Y. et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965 (2017).
Gao, M. J. et al. Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21, 54–71 (2009).
Liu, H. et al. Distant eQTLs and non-coding sequences play critical roles in regulating gene expression and quantitative trait variation in maize. Mol. Plant 10, 414–426 (2017).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Felsenstein, J. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5, 164–166 (1989).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Hu, J. et al. NextDenovo: an efficient error correction and accurate assembly tool for noisy long reads. Genome Biol. 25, 107 (2024).
Hu, J., Fan, J., Sun, Z. & Liu, S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics 36, 2253–2255 (2020).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Alonge, M. et al. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol. 23, 258 (2022).
Marcais, G. et al. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 14, e1005944 (2018).
Ou, S. & Jiang, N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176, 1410–1422 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Liu, R. & Dickerson, J. Strawberry: fast and accurate genome-guided transcript reconstruction and quantification from RNA-Seq. PLoS Comput. Biol. 13, e1005851 (2017).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Song, L., Sabunciyan, S. & Florea, L. CLASS2: accurate and efficient splice variant annotation from RNA-seq reads. Nucleic Acids Res. 44, e98 (2016).
Venturini, L., Caim, S., Kaithakottil, G. G., Mapleson, D. L. & Swarbreck, D. Leveraging multiple transcriptome assembly methods for improved gene structure annotation. Gigascience 7, giy093 (2018).
Mapleson, D., Venturini, L., Kaithakottil, G. & Swarbreck, D. Efficient and accurate detection of splice junctions from RNA-seq with Portcullis. Gigascience 7, giy131 (2018).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinform. 10, 421 (2009).
Bruna, T., Hoff, K. J., Lomsadze, A., Stanke, M. & Borodovsky, M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinform. 3, lqaa108 (2021).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 25, 4–10 (2009).
Dainat, J. & Pucholt, D. H. AGAT: another Gff analysis toolkit to handle annotations in any GTF. v.0.6.0. Zenodo https://doi.org/10.5281/zenodo.4637977 (2021).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003).
Soderlund, C. et al. Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet. 5, e1000740 (2009).
Wang, B. et al. A comparative transcriptional landscape of maize and sorghum obtained by single-molecule sequencing. Genome Res. 28, 921–932 (2018).
Zhang, R. G. et al. TEsorter: an accurate and fast method to classify LTR-retrotransposons in plant genomes. Hortic. Res. 9, uhac017 (2022).
Campbell, M. S. et al. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164, 513–524 (2014).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Chen, M. M., Lin, H., Chiang, L. M., Childers, C. P. & Poelchau, M. F. The GFF3toolkit: QC and merge pipeline for genome annotation. Methods Mol. Biol. 1858, 75–87 (2019).
Blum, M. et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354 (2021).
Olson, A. J. & Ware, D. Ranked choice voting for representative transcripts with TRaCE. Bioinformatics 38, 261–264 (2021).
Ou, S. et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20, 275 (2019).
Wenke, T. et al. Targeted identification of short interspersed nuclear element families shows their widespread existence and extreme heterogeneity in plant genomes. Plant Cell 23, 3117–3128 (2011).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Goel, M., Sun, H., Jiao, W. B. & Schneeberger, K. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20, 277 (2019).
Kronenberg, Z. N. et al. High-resolution comparative analysis of great ape genomes. Science 360, eaar6343 (2018).
Sedlazeck, F. J. et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018).
Jiang, T. et al. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 21, 189 (2020).
Hickey, G. et al. Genotyping structural variants in pangenome graphs using the vg toolkit. Genome Biol. 21, 35 (2020).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Chen, S. et al. Paragraph: a graph-based structural variant genotyper for short-read sequence data. Genome Biol. 20, 291 (2019).
Speed, D., Holmes, J. & Balding, D. J. Evaluating and improving heritability models using summary statistics. Nat. Genet. 52, 458–462 (2020).
Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–824 (2012).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Chen, S., Songkumarn, P., Liu, J. & Wang, G. L. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121 (2009).
Xing, H. L. et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 (2014).
Byeon, B. et al. The ATP-dependent chromatin remodeling enzyme Fun30 represses transcription by sliding promoter-proximal nucleosomes. J. Biol. Chem. 288, 23182–23193 (2013).
Yang, S. Supplementary data of the maize pan-genome work (1.0). Zenodo https://doi.org/10.5281/zenodo.16576184 (2025).
Yang, S. GWAS results related to the maize pan-genome work (1.0). Zenodo https://doi.org/10.5281/zenodo.17138577 (2025).
Yang, S. Maize pan-genome related scripts and pipelines (1.0). Zenodo https://doi.org/10.5281/zenodo.16631382 (2025).
Acknowledgements
We thank T. Tian (University of Science and Technology Beijing), Z. Zhang (China Agricultural University) and H. He (Fujian Agriculture and Forestry University) for valuable discussions, and S. Wang and Y. Liu (Instrumental platform of state key laboratory of plant environmental resilience, China Agricultural University, Beijing) for the help on scanning electron microscopy and confocal imaging. This research was supported by the National Key Research and Development Program of China (grant number 2023YFF1001300), the National Natural Science Foundation of China (grant numbers 32430010 and 32272024), the Chinese Universities Scientific Fund (grant numbers 2025TC135 and 2025TC148), Beijing Outstanding Young Scientist Program (grant number BJJWZYJH01201910019026) and the China Postdoctoral Science Foundation (grant number 2019M660867).
Author information
Authors and Affiliations
Contributions
F.Q. and S.Y. designed and supervised the study and revised the paper. S.Y. and Y.W. performed the pangenome study and gene association analysis. Q.H., M.W., S.W., X.F., C.Z., S.L. and Z.Y. performed experiments for gene cloning and collected phenotypic data in the fields. J.C. contributed to transgenic maize generations. N.Y., J.Y. and X.Y. provided maize materials and valuable discussions and edited the paper. All the authors read and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
Two patent applications related to this work have been submitted by F.Q., S.Y., Q.H. and M.W. The other authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Klaus Mayer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Drought resistance phenotypes analysis in pan-genome and population level.
a, Representative plant photographs of the 23 NAM founders16, excluding B73 (presented in Tian et al.25), CML247 and CML277 (lacking sufficient seeds), grown under WW and WS2 conditions in fields. The ears are harvested from the plants grown under WW, WS1 and WS2 conditions. They are ordered according to the average yield under WS1 and WS2. The size of bubble corresponds to the average yield and days of ASI under WS1 and WS2 conditions, and seedling survival rate (SR) after drought stress. b, Integrative illustration of multiple drought resistance phenotypes of 50 germplasms. The 50 germplasms include 25 newly assembled germplasms in this study, 26 NAM founders16, Mo1727 and CIMBL5525, excluding SY1032, CML247 and CML277 (lacking sufficient seeds). Plant drought resistance at the seedling stage is scored according to the survival rate after drought. All the germplasms are classified into three categories: ‘Resistant’, ‘Intermediate’ and ‘Sensitive’ as Fig. 1c. ‘WW yield’, grain yield under WW; ‘WS yield’, (yield_WS1+yield_WS2)/2; ‘dASI’, (ASI_WS1+ASI_WS2)/2 − (ASI_WW). The stacked bar value (left y-axis) represents WW yield (light blue) and WS yield (bright blue) for each germplasm. Scatter points (right y-axis) show the dASI for each germplasm. Red inverted triangles indicate six drought-resistant germplasms referring to three criteria: 1) resistance or intermediate resistance at the seedling stage; 2) yield > 25g (above the cyan dotted line) under WS; 3) dASI < 2 days (below the red dotted line). c, Statistic of ASI under WW and WS conditions. Statistical significance was determined by paired t-test. d, e, Correlation between ASI and yield per plant (d), seedling SR and yield per plant (e) under WW and WS conditions. Pearson correlation coefficient (r) is used to evaluate a linear correlation between two traits. Statistical significance was determined by a two-sided t-test. In c–e, the phenotypes are obtained from 228 temperate germplasms9.
Extended Data Fig. 2 Plant survival rate assay for drought resistance at seedling stage for NAM founders.
The seedling SR of each germplasm is compared with B73, respectively. Fifteen plants of each germplasm are compared in each pot, with at least two replicates. Representative photographs are taken before and after drought treatment.
Extended Data Fig. 3 Pipeline of de novo assembly for 25 genomes.
Approximately 98× genomic DNA long-read sequencing data generated by Oxford Nanopore Technology (ONT) were initially corrected by NextDenovo54. Then, the ONT reads and about 60× genomic DNA short-read sequencing data generated by Illumina NovaSeq 6000 were employed to polish draft contigs. RagTag58 was utilized to address inter-chromosomal assembly errors and construct chromosome-scale pseudomolecules (See Methods).
Extended Data Fig. 4 Pipeline of gene and repeat sequence annotation.
a, Gene annotation pipeline. Both evidence-based and ab initio gene prediction methods were used for each genome. For the evidence-based method, transcript assembly programs were used for transcript prediction based on RNA-seq data of five tissues (See Methods) and Mikado67 was further employed to select the best transcripts. For ab initio gene prediction, the mapped RNA-seq reads and protein sequences generated by evidence-based method were used as inputs of BRAKER271 for gene predictions. The non-redundant gene models from both methods were further refined by PASA274 based on maize Iso-seq76 and Expressed Sequence Tags (ESTs) from GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The final gene models were generated after post-processing. b, Repeat sequence annotation pipeline. For long terminal repeat (LTR), terminal inverted repeat (TIR), and Helitron annotation, novel transposable elements (TEs) for each genome were identified using RepeatMasker72 and EDTA83 by comparing with a known TE library (METC, https://github.com/oushujun/MTEC). After merging and removing redundant novel TEs from 25 novel assembly genomes, a non-redundant novel library was created. This novel TE library was further aggregated with the MTEC library to form the final Pan-TE library. LTRs, TIRs, and helitrons were finally annotated based on the Pan-TE library. Besides, short interspersed nuclear elements (SINEs) were identified by SineFinder84, while long interspersed nuclear elements (LINEs) and non-TE repeats were directly annotated by RepeatMasker and EDTA.
Extended Data Fig. 5 Pan-gene and Pan-SV analyses of 56 maize genomes.
a, b Modeling the size of maize pan-genes and core-genes (a), pan-gene families and core-gene families (b) when additional genomes are incorporated into the maize pan-genome. Genomes were sampled as 56 random combinations of each given number of genomes. Mean values are displayed with error bars representing ± SD. c, Observed (arrow) and expected (density) distribution of genes belonging to core and softcore gene families. The expected distribution is the proportion of genes belonging to core and softcore gene families by randomly sampling 1,344 protein-coding genes for 10,000 times. The statistical significance is determined by permutation test. d, Accumulation of different types of SVs, including insertion (INS), deletion (DEL), duplication (DUP), translocation (TRA), and inversion (INV), with the increase of genome numbers. The height of the stacked chart represents the total number of SVs, while the colored sections show the number of different types of SV. e, Upset plot showing the number of SVs identified by different methods. High-quality SVs refer to those identified by at least two methods. The horizontal bars represent the total number of SVs identified by each method. Vertical bars display the number of SVs identified by one or multiple methods, as indicated by the black dots below the x-axis. Black dots indicate the methods for the SVs identification. f, Distribution of pan-SV position, referring to the gene position in B73 (v5). ‘UTR’, untranslated region. ‘CDS’, coding region sequence. g, Proportion of pan-SVs of different lengths. h, Root gravitropism assay for B73 and CML333. Red arrow indicates the direction of gravity. The inclination angle of the root with the horizontal line was measured, 12 hours after rotation the petri dishes 90° to the right. Numbers above the x-axis indicate the sample size, and the statistical significance is determined by a two-sided t-test.
Extended Data Fig. 6 Pipeline for pangenome structural variant (pan-SV) identification.
Initial SV identification was based on a maize pan-genome comprising 56 high-quality assembled genomes. The Nanopore/PacBio long-read sequences of each germplasm were mapped to the B73 (v5) genome sequence and SVs were identified by Sniffles88 and CuteSV89. Meanwhile, SyRI86 and Smartie-sv87 were employed to identify SVs through sequence alignment of each assembled genome sequence to that of B73 (v5). SVs identified in NAM founders16 were merged into the final non-redundant SV dataset using Jasmine17.
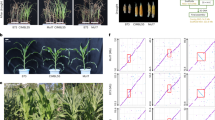
Extended Data Fig. 7 ZmUGE2 contributes to seedling drought resistance.
a–c, Manhattan plot of the GWAS identifying the genetic loci associated with the ZmUGE2 expression levels under WW (a), WS1 (b) and WS2 (c) conditions. Variants located within ZmUGE2 and the 5-kb flanking sequence are indicated by red dots. d, Comparison of gene expression levels of ZmUGE2-Hap1 and ZmUGE2-Hap2 under WW and WS2 conditions. e, Comparison of gene expression levels of ZmUGE2 (left panel) and the survival rates (right panel) of the germplasms carrying different genotypes of ZmUGE2 based on the genotype of Indel no. 7434. ‘+’ indicates the presence of the 9-bp insertion; ‘−’ indicates its absence. f, Left panel: RT-qPCR analysis of relative ZmUGE2 transcript levels in WT and ZmUGE2-OE lines, normalized to the internal control gene ZmUBI. Mean values are displayed with error bars representing ± SD from three independent biological replicates. Right panel: Western blot analysis of the ZmUGE2-GFP protein in the ZmUGE2-OE plants, with actin used as a loading control. Molecular weight markers are shown on the right. g, Schematic diagram of the CRISPR-targeted knockout genotype of zmuge2-KO lines. The gRNA target sequences and edits are indicated below the gene diagram. h, Water loss rate (%) of detached leaves at the indicated time points. For each genotype, four detached leaves are placed on a clean bench to dehydrate, and their weights are recorded periodically over an 8-hour period. Data represent the mean ± SD, based on three replicated experiments. Asterisks (*p < 0.05) indicate a significant difference between the WT and transgenic plants. i, Comparison of plant height between WT and ZmUGE2-OE plants. Numbers above the x-axis indicate the number of plants for each genotype. Numbers above the x-axis represent the number of germplasms for each genotype in d and e. In d–f, h and i, statistical significance is determined by a two-sided t-test.
Extended Data Fig. 8 ZmSIL2 plays a negative role in maize seedling drought resistance.
a-c, Manhattan plot of the GWAS identifying the genetic loci associated with ZmSIL2 expression levels under WW (a), WS1 (b) and WS2 (c) conditions. Variants located within ZmSIL2 and the 5-kb flanking sequence are indicated by red dots. d, Comparison of gene expression levels of ZmSIL2-Hap1 and ZmSIL2-Hap2 under WW and WS2 conditions. e, Comparison of gene expression levels of ZmSIL2 (left panel) and the survival rates (right panel) of the germplasms carrying different genotypes of ZmSIL2 based on SV no. 2606. ‘+’ indicates the presence of the 715-bp insertion; ‘−’ indicates its absence. f, Expression level of ZmSIL2 in WT and ZmSIL2-OE lines. Mean ± SD from three independent biological replicates. g, Schematic diagram of the CRISPR-targeted knockout genotype of ZmSIL2. The trihelical DNA-binding domain is highlighted, and the predicted α-helix regions are shown in cyan. h, Subcellular localization of ZmSIL2-GFP. Confocal microscopy images the expressed ZmSIL2-GFP protein in maize leaf protoplasts. Scale bar, 5 μm. i, Western blot analysis of the samples used for the ZmSIL2-GFP ChIP-seq analysis. ‘WT’, the samples of WT are prepared parallelly as negative controls. ‘Anti-H3’ indicates the nuclear fraction. j, Gene expression analysis of four ZmSIL2-regulated genes. For each gene, the gene structure is shown in the upper panel, and the RNA-seq reads graphs of WT and zmsil2-KO1 samples under WW and WS conditions are shown in the four tracks in the middle panel. The bottom panel shows RT-qPCR confirmation of the altered gene expression in ZmSIL2-OE and zmsil2-KO plants. Data are presented as mean values ± SD. In d and e, numbers above the x-axis represent the number of germplasms for each genotype. In h and i, the micrographs and immunoblot are the representative results from at least two independent biological replicates. In d–f and j, statistical significance is determined by a two-sided t-test.
Extended Data Fig. 9 ZmASI3 contributes to maize drought resistance at the flowering stage.
a, The putative SNF2 helicase domains (ZmASI3-1267 aa) encoded by T01 and the truncated one (ZmASI3-1151) encoded by T02 are fused with Glutathione S-transferase (GST) for protein expression and purification in E. coli, respectively. b, Protein gel electrophoresis for the purified GST, GST-ZmASI3-1267, and GST-ZmASI3-1151 proteins. Molecular weight markers are indicated on the right side. c, In vitro ATPase activity assay. GST, GST-ZmASI3-1267, and GST-ZmASI3-1151 proteins were used in the assay. ‘+’ indicates 0.2 μg of protein, while ‘++’ indicates 0.4 μg. Mean values are displayed with error bars representing ± SD from three independent biological replicates. d, Schematic illustration for the in vitro nucleosome remodeling assay. The previously inaccessible Dpn II site (yellow) became exposed due to localized nucleosome sliding. e, Nucleosome remodeling assay. The GST-ZmASI3-1267 and GST-ZmASI3-1151 proteins are incubated with the pre-assembled nucleosome over the indicated time periods. Bands near 225 bp represent intact nucleosomal DNA, while bands near 200 bp indicate fragment mobility cleaved by Dpn II after nucleosome remodeling, indicated by the red asterisk. GST protein was used as a negative control. The gel electrophoresis is the representative result from two independent biological replicates. f, Presence and absence of SV1-5 within intron 5 of ZmASI3 among different haplotypes. Cyan indicates the presence of an SV, while gray indicates its absence. g, Schematic diagram of the CRISPR-targeted knockout genotype of ZmASI3. Violin plots of days to anthesis (DTA) (h) and days to silking (DTS) (i) in WT and zmasi3-KO plants under WW and WS field conditions. In h and i, Numbers above the x-axis represent the number of plants for each genotype, and statistical significance is determined by a two-sided t-test.
Supplementary information
Supplementary Information
Supplementary Note, Tables 1–16 and refs. 1–13.
Supplementary Tables
Supplementary Tables 1–16.
Source data
Figs. 1–5 and Extended Data Figs. 1, 5 and 7–9
Statistical source data for Figs. 1–5 and Extended Data Figs. 1, 5 and 7–9.
Fig. 3g and Extended Data Figs. 7–9
Unprocessed western blots and gels for Fig. 3g and Extended Data Figs. 7f, 8i and 9b,e.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, S., Wang, Y., Huang, Q. et al. A pangenome of maize provides genetic insights into drought resistance. Nat Genet 57, 2831–2841 (2025). https://doi.org/10.1038/s41588-025-02378-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-025-02378-w