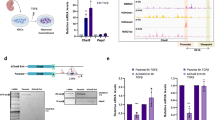
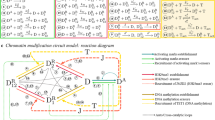
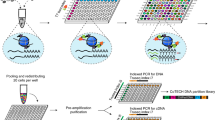
Abstract
Enhancer–promoter (E-P) interactions regulate transcription during cell fate determination. However, the regulatory mechanisms underlying E-P interactions have remained elusive. Here we present a chromatin-interaction-based proteomic approach, LoopID, to profile proteins (termed the looposome) at certain E-P anchors. We find that histone demethylase JMJD2, a key looposome component, can regulate E-P interactions and the looposome in a catalytic-independent manner through formation of biomolecular condensates. Furthermore, we introduce a system to engineer E-P interactions by assembling JMJD2 condensates at certain genomic loci, enabling construction of cell-type-specific E-P interactions to promote cellular reprogramming into pluripotent or two-cell-like cells. Our findings reveal a noncanonical function of a histone demethylase in regulation of chromatin organization and provide a strategy to regulate cell fate transitions through E-P interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All omics data generated in this study are summarized in Supplementary Table 8. HiChIP (mES cell H3K27ac), AQuA-HiChIP (H3K27ac), Micro-C (mES cell), 4C–seq (mES cell E-P loop of Zscan4c), ChIP–seq and Cut&Tag (mES cell H3K27ac, H3K36me3, H3K4me3, H3K9me2, H3K9me3, JMJD2A, JMJD2C, SadCas9, SpdCas9, YY1) are available through the Gene Expression Omnibus under accession GSE232848. The proteomics data generated in this study have been deposited in PRIDE under accession number PXD069064. Public ChIP–seq data from mES cells: BRD4 (Sabari et al.7, GSE112808); CBX2, EZH2 and RING1B (Deaton et al.56, GSE78899); CTCF, H3K27ac, H3K4me1 and H3K4me3 (Shen et al.50, GSE29218); H3K27ac and p300 (Chronis et al.57, GSE90893); H3K9me3 (Cho et al.58, GSE106176); HDAC1, HDAC2 and LSD1 (Whyte et al.59, GSE27841); HP1 and SMYD5 (Kidder et al.60, GSE94033); JARID1B (Schmitz et al.61, GSE31966); Jmjd2a/b/c-TKO (long-term) H3K9me3 (Pedersen et al.22, GSE64254); JMJD2B and JMJD2C (Das et al.36, GSE43231); JMJD3 (Banaszynski et al.62, GSE42152); MLL2 and SUZ12 (Mas et al.63, GSE99530); MLL3/4 (Dorighi et al.64, GSE98063); MLL4 (Cao et al.65, GSE99022); OCT4 (Whyte et al.66, GSE44286); PRDM4 (Bogani et al.67, GSE48372); SET1A (Sze et al.68, GSE98988); SETDB1 (Bilodeau et al.69, GSE18371); SUV39H1 and SUV39H2 (Bulut-Karslioglu et al.70, GSE57092); TET1 (Williams et al.71, GSE24841); YY1 (Weintraub et al.9, GSE99518); USF1 and WDR5 (Scelfo et al.72, GSE122715); UTX (Wang et al.73, GSE103180). Public ChIP–seq data from two-cell embryos: H3K27ac and H3K4me3 (Dahl et al.74, GSE72784). Public ChIP–seq data from 2CLCs: H3K27ac and H3K4me3 (Zhu et al.46, GSE159623). Public 3D genome data from mES cells: H3K27ac HiChIP (Mumbach et al.14, GSE101498); from two-cell embryos: Hi-C (Du et al.75, GSE82185); from 2CLCs: Hi-C (Zhu et al.46, GSE159623). Public RNA-seq data from from 2CLCs (Zhu et al.46, GSE159623), ciTotiSCs (Hu et al.76, GSE185000); mES cells, D-EPSCs and L-EPSCs (Posfai et al.77, GSE145609); two-cell embryos (Zhang et al.78, GSE71434); TLSCs (Yang et al.54, GSE166204); bulk RNA-seq of mouse embryos (Wu et al.79, GSE66390); and single-cell RNA-seq from mouse embryos (Fan et al.80, GSE53386), totipotent blastomere-like cells (Shen et al.81, GSE168728) and totipotent-like stem cells (Xu et al.82, GSE183522). Source data are provided with this paper.
Code availability
Software and algorithms used in this study were as follows: scripts written in-house for this study (Calculation of maintaining score, https://github.com/XinyiLiu671/LoopID), 10X Genomics Cell Ranger v.7.1.0 (Zheng et al.83, https://www.10xgenomics.com), Basic4Cseq v.1.38.0 (Walter et al.84, https://bioconductor.org/packages/release/bioc/html/Basic4Cseq.html), bedtools v.2.26.0 (Quinlan et al.85, https://github.com/arq5x/bedtools), BioGPS (Wu et al.86, http://biogps.org/#goto=welcome), Bowtie2 v.2.3.0 (Langmead et al.87, https://github.com/arq5x/bedtools), Cas-OFFinder (Bae et al.88, http://www.rgenome.net/cas-offinder/), CRISPick (https://portals.broadinstitute.org/gppx/crispick/public), crisprScore v.1.10.0, (Hoberecht et al.89, https://www.bioconductor.org/packages/release/bioc/html/crisprScore.html), Cytoscape v.3.9.1 (Shannon et al.90, https://cytoscape.org), DAVID (Sherman et al.55, https://david.ncifcrf.gov/tools.jsp), DeepTools v.3.5.1 (Ramírez et al.91, https://deeptools.readthedocs.io/en/develop/content/list_of_tools.html), diffloop v.1.14.0 (Lareau et al.92, http://bioconductor.org/packages/release/bioc/html/diffloop.html), EdgeR v.3.26.5 (Robinson et al.93, https://bioconductor.org/packages/release/bioc/html/edgeR.html), Fastp v.0.23.2 (Chen et al.94, https://github.com/OpenGene/fastp), FourCSeq v.1.2.0 (Klein et al.95, https://bioconductor.riken.jp/packages/3.1/bioc/html/FourCSeq.html), GENOVA v.0.95 (van der Weide et al.96, https://github.com/robinweide/GENOVA), ggplot2 v.3.4.3 (Wickham97, https://ggplot2.tidyverse.org), ggpubr v.0.6.0 (Kassambara98, https://cran.r-project.org/web/packages/ggpubr/index.html), GSEA v.4.0.3 (Subramanian et al.99, https://www.gsea-msigdb.org/gsea/index.jsp), HiCExplore v.3.7.1 (Wolff et al.100, https://hicexplorer.readthedocs.io/en/latest/index.html), hichipper v.0.7.5 (Lareau et al.101, https://github.com/aryeelab/hichipper), HiC-Pro v.2.10.0 (Servant et al.102, https://github.com/nservant/HiC-Pro), HOMER v.4.11 (Heinz et al.103, http://homer.ucsd.edu/homer), HTSeq-count v.0.11.2 (Anders et al.104, https://github.com/simon-anders/htseq), IGV v.2.4.10 (Thorvaldsdottir et al.105, http://software.broadinstitute.org/software/igv), ImageJ (Schneider et al.106, https://imagej.nih.gov), Imaris (Bitplane, https://imaris.oxinst.com), Juicebox v.1.9.8 (Robinson et al.107, https://www.aidenlab.org/juicebox), Macs2 v.2.1.2 (Zhang et al.108, https://github.com/taoliu/MACS), Mouse Reference Genome, NCBI build 37, NCBI37/mm9 (Genome Reference Consortium, https://www.ncbi.nlm.nih.gov/grc/mouse), PANTHER (Thomas et al.109, https://www.pantherdb.org), Perl v.5.34.0 (Wall et al.110, https://www.perl.org), SAINTexpress v.3.3 (Teo et al.17, https://saint-apms.sourceforge.net/Main.html).
References
Heinz, S. et al. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Bonev, B. et al. Organization and function of the 3D genome. Nat. Rev. Genet. 17, 772 (2016).
Krijger, P. H. L. et al. Regulation of disease-associated gene expression in the 3D genome. Nat. Rev. Mol. Cell Biol. 17, 771–782 (2016).
Hnisz, D. et al. A phase separation model for transcriptional control. Cell 169, 13–23 (2017).
Boija, A. et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855 (2018).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, aar3958 (2018).
Wang, W. et al. A histidine cluster determines YY1-compartmentalized coactivators and chromatin elements in phase-separated enhancer clusters. Nucleic Acids Res. 50, 4917–4937 (2022).
Weintraub, A. S. et al. YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573–1588 (2017).
Ahn, J. H. et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 595, 591–595 (2021).
Cho, K. F. et al. Split-TurboID enables contact-dependent proximity labeling in cells. Proc. Natl Acad. Sci. USA 117, 12143–12154 (2020).
Costa, Y. et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370–374 (2013).
Ding, J. et al. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 22, 155–167 (2012).
Mumbach, M. R. et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49, 1602–1612 (2017).
Peng, T. et al. STARR-seq identifies active, chromatin-masked, and dormant enhancers in pluripotent mouse embryonic stem cells. Genome Biol. 21, 243 (2020).
Chen, H. et al. Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet. 50, 1296–1303 (2018).
Teo, G. et al. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics 100, 37–43 (2014).
Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018).
Ji, X. et al. Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc. Natl Acad. Sci. USA 112, 3841–3846 (2015).
Hammal, F. et al. ReMap 2022: a database of human, mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res. 50, D316–D325 (2022).
Tsukada, Y. et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006).
Pedersen, M. T. et al. Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J. 35, 1550–1564 (2016).
Mumbach, M. R. et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016).
Gryder, B. E. et al. Measurement of differential chromatin interactions with absolute quantification of architecture (AQuA-HiChIP). Nat. Protoc. 15, 1209–1236 (2020).
Krietenstein, N. et al. Ultrastructural details of mammalian chromosome architecture. Mol. Cell 78, 554–565 (2020).
Hsieh, T. S. et al. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell 78, 539–553 (2020).
Alberti, S. et al. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019).
Tomaz, R. A. et al. Jmjd2c facilitates the assembly of essential enhancer-protein complexes at the onset of embryonic stem cell differentiation. Development 144, 567–579 (2017).
Bracha, D. et al. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175, 1467–1480 (2018).
Jiang, Y. et al. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat. Genet. 49, 1239–1250 (2017).
Yan, J. et al. Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res. 28, 204–220 (2018).
Yan, Z. et al. G9a/GLP-sensitivity of H3K9me2 demarcates two types of genomic compartments. Genomics Proteomics Bioinformatics 18, 359–370 (2020).
Montavon, T. et al. Complete loss of H3K9 methylation dissolves mouse heterochromatin organization. Nat. Commun. 12, 4359 (2021).
Morgan, M. A. J. et al. Epigenetic moonlighting: catalytic-independent functions of histone modifiers in regulating transcription. Sci. Adv. 9, eadg6593 (2023).
Hsieh, T. S. et al. Enhancer-promoter interactions and transcription are largely maintained upon acute loss of CTCF, cohesin, WAPL or YY1. Nat. Genet. 54, 1919–1932 (2022).
Das, P. P. et al. Distinct and combinatorial functions of Jmjd2b/Kdm4b and Jmjd2c/Kdm4c in mouse embryonic stem cell identity. Mol. Cell 53, 32–48 (2014).
Wang, J. et al. YY1 positively regulates transcription by targeting promoters and super-enhancers through the BAF complex in embryonic stem sells. Stem Cell Rep. 10, 1324–1339 (2018).
Olley, G. et al. BRD4 interacts with NIPBL and BRD4 is mutated in a Cornelia de Lange-like syndrome. Nat. Genet. 50, 329–332 (2018).
Schoenfelder, S. et al. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019).
Morgan, S. L. et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun. 8, 15993 (2017).
Le, R. et al. Dcaf11 activates Zscan4-mediated alternative telomere lengthening in early embryos and embryonic stem cells. Cell Stem Cell 28, 732–747 (2021).
Chen, M. et al. Chromatin architecture reorganization in murine somatic cell nuclear transfer embryos. Nat. Commun. 11, 1813 (2020).
Wang, J. et al. Phase separation of OCT4 controls TAD reorganization to promote cell fate transitions. Cell Stem Cell 28, 1868–1883 (2021).
Hu, Z. et al. Maternal factor NELFA drives a 2C-like state in mouse embryonic stem cells. Nat. Cell Biol. 22, 175–186 (2020).
Amano, T. et al. Zscan4 restores the developmental potency of embryonic stem cells. Nat. Commun. 4, 1966 (2013).
Zhu, Y. et al. Relaxed 3D genome conformation facilitates the pluripotent to totipotent-like state transition in embryonic stem cells. Nucleic Acids Res. 49, 12167–12177 (2021).
Yang, F. et al. DUX-miR-344-ZMYM2-mediated activation of MERVL LTRs induces a totipotent 2C-like state. Cell Stem Cell 26, 234–250 (2020).
Kadauke, S. et al. Chromatin loops in gene regulation. Biochim. Biophys. Acta 1789, 17–25 (2009).
Anania, C. et al. Order and disorder: abnormal 3D chromatin organization in human disease. Brief. Funct. Genomics 19, 128–138 (2020).
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012).
Wang, J. et al. A comprehensive atlas of epigenetic regulators reveals tissue-specific epigenetic regulation patterns. Epigenetics 18, 2139067 (2023).
Hu, H. et al. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 47, D33–D38 (2019).
Pedersen, M. T. et al. The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Mol. Cell. Biol. 34, 1031–1045 (2014).
Yang, M. et al. Chemical-induced chromatin remodeling reprograms mouse ESCs to totipotent-like stem cells. Cell Stem Cell 29, 400–418 (2022).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 50, W216–W221 (2022).
Deaton, A. M. et al. Enhancer regions show high histone H3.3 turnover that changes during differentiation. eLife 5, e15316 (2016).
Chronis, C. et al. Cooperative binding of transcription factors orchestrates reprogramming. Cell 168, 442–459 (2017).
Cho, S. W. et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 173, 1398–1412 (2018).
Whyte, W. A. et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225 (2012).
Kidder, B. L. et al. SMYD5 regulates H4K20me3-marked heterochromatin to safeguard ES cell self-renewal and prevent spurious differentiation. Epigenetics Chromatin 10, 8 (2017).
Schmitz, S. U. et al. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J. 30, 4586–4600 (2011).
Banaszynski, L. A. et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013).
Mas, G. et al. Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat. Genet. 50, 1452–1462 (2018).
Dorighi, K. M. et al. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol. Cell 66, 568–576 (2017).
Cao, K. et al. An Mll4/COMPASS-Lsd1 epigenetic axis governs enhancer function and pluripotency transition in embryonic stem cells. Sci. Adv. 4, eaap8747 (2018).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Bogani, D. et al. The PR/SET domain zinc finger protein Prdm4 regulates gene expression in embryonic stem cells but plays a nonessential role in the developing mouse embryo. Mol. Cell. Biol. 33, 3936–3950 (2013).
Sze, C. C. et al. Histone H3K4 methylation-dependent and -independent functions of Set1A/COMPASS in embryonic stem cell self-renewal and differentiation. Genes Dev. 31, 1732–1737 (2017).
Bilodeau, S. et al. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 23, 2484–2489 (2009).
Bulut-Karslioglu, A. et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell 55, 277–290 (2014).
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011).
Scelfo, A. et al. Functional landscape of PCGF proteins reveals both RING1A/B-dependent-and RING1A/B-independent-specific activities. Mol. Cell 74, 1037–1052 (2019).
Wang, A. H. et al. The elongation factor Spt6 maintains ESC pluripotency by controlling super-enhancers and counteracting Polycomb proteins. Mol. Cell 68, 398–413 (2017).
Dahl, J. A. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016).
Du, Z. et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547, 232–235 (2017).
Hu, Y. et al. Induction of mouse totipotent stem cells by a defined chemical cocktail. Nature 617, 792–797 (2023).
Posfai, E. et al. Evaluating totipotency using criteria of increasing stringency. Nat. Cell Biol. 23, 49–60 (2021).
Zhang, B. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016).
Wu, J. et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016).
Fan, X. et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148 (2015).
Shen, H. et al. Mouse totipotent stem cells captured and maintained through spliceosomal repression. Cell 184, 2843–2859 (2021).
Xu, Y. et al. Derivation of totipotent-like stem cells with blastocyst-like structure forming potential. Cell Res. 32, 513–529 (2022).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Walter, C. et al. Basic4Cseq: an R/Bioconductor package for analyzing 4C-seq data. Bioinformatics 30, 3268–3269 (2014).
Quinlan, A. R. et al. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Wu, C. et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130 (2009).
Langmead, B. et al. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Bae, S. et al. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Hoberecht, L. et al. A comprehensive Bioconductor ecosystem for the design of CRISPR guide RNAs across nucleases and technologies. Nat. Commun. 13, 6568 (2022).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Lareau, C. A. et al. diffloop: a computational framework for identifying and analyzing differential DNA loops from sequencing data. Bioinformatics 34, 672–674 (2018).
Robinson, M. D. et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Chen, S. et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Klein, F. A. et al. FourCSeq: analysis of 4C sequencing data. Bioinformatics 31, 3085–3091 (2015).
van der Weide, R. H. et al. Hi-C analyses with GENOVA: a case study with cohesin variants. NAR Genom. Bioinform. 3, lqab040 (2021).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016).
Kassambara, A. ggpubr: ‘ggplot2’ based publication ready plots. https://cran.r-project.org/web/packages/ggpubr/index.html (2023).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Wolff, J. et al. Galaxy HiCExplorer 3: a web server for reproducible Hi-C, capture Hi-C and single-cell Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 48, W177–W184 (2020).
Lareau, C. A. et al. hichipper: a preprocessing pipeline for calling DNA loops from HiChIP data. Nat. Methods 15, 155–156 (2018).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Anders, S. et al. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Thorvaldsdottir, H. et al. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Schneider, C. A. et al. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Robinson, J. T. et al. Juicebox.js provides a cloud-based visualization system for Hi-C data. Cell Syst. 6, 256–258 (2018).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Thomas, P. D. et al. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 31, 8–22 (2022).
Wall, L., Christiansen, T. & Orwant, J. Programming Perl (O’Reilly Media, 2000).
Acknowledgements
We thank K. Helin and K. Agger (University of Copenhagen, Denmark) for help with JMJD2 cell lines, and R. A. Young (Whitehead Institute for Biomedical Research, USA) for pET-mEGFP plasmids. This work was funded by the National Key Research and Development Program of China (2023YFA1800900) (J.J.D.); National Key Research and Development Program of China (2024YFA1106900) (J.J.D.); National Science Foundation for Distinguished Young Scholars of China (32425022) (J.J.D.); National Natural Science Foundation of China (32170798 and 32430031) (J.J.D.); Guangdong Innovative and Entrepreneurial Research Team Program (2016ZT06S029) (J.J.D.); National Natural Science Foundation of China (32300669) (S.S.J.); Sun Yat-sen University Fundamental Research Funds for Colleges and Universities Young Teachers Cultivation Project (23ptpy89) (S.S.J.); Natural Science Foundation of Guangdong Province (2024A1515011174) (S.S.J.); National Natural Science Foundation of China (32400659) (X.Y.L.); China Postdoctoral Science Foundation (2023M744082) (X.Y.L.); Natural Science Foundation of Guangdong Province (2023A1515010148) (J.S.); National Natural Science Foundation of China (32400561) (J.S.); Guangdong Basic and Applied Basic Research Foundation (2025B1515020011) (L.L.F.); Guangzhou Science and Technology Program Project, China (No.2025A04J5154) (L.L.F.); National Natural Science Foundation of China (82304746) (L.L.F.); National Natural Science Foundation of China (323B2026) (J.L.Q.); and NIH/NHGRI (1R35HG010717-01) (L.P.); Rappaport MGH Research Scholar Award 2024-2029 (L.P.).
Author information
Authors and Affiliations
Contributions
S.J., X.L. and J.D. designed the study; S.J., X.L. and J.D. discussed the results and wrote the manuscript with input from all other authors; X.L. performed bioinformatic analyses and visualizations; X.H., J.T., Z. Zhang. and L.M. conducted experiments on biomolecular condensates; X.Z., X.L., H. Lin., X.H. and Z. Zhang performed the LoopID, HiChIP and ChIP experiments; Q.T., Z.W., Z. Zhou, L.Z., H. Li. and J.Q. constructed plasmids and performed transfections; P.P.D. and L.P. provided the ChIP–seq data for JMJD2A and KDM4A; J.L. and Z.D. prepared nucleosome arrays modified with H3K4me3; M.Y. generated the chimeras; and J.S., J.W., H.W., D.-f.H., J.B., L.F., W.C., X.X., J.C.W., D.G. and L.W. provided guidance for the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Alessio Zippo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–8, Figs. 1–8 (including all associated legends), unprocessed gels or blots for supplementary figures, and references.
Supplementary Table 1
Supplementary Tables 1–8.
Supplementary Data 1
Statistical source data for supplementary figures.
Source data
Source Data Fig. 1
Unprocessed western blots and/or gels for Fig. 1.
Source Data Fig. 2
Statistical source data for Fig. 2.
Source Data Fig. 3
Statistical source data for Fig. 3.
Source Data Fig. 4
Statistical source data for Fig. 4.
Source Data Fig. 5
Unprocessed western blots and/or gels for Fig. 5.
Source Data Fig. 5
Statistical source data for Fig. 5.
Source Data Fig. 6
Unprocessed western blots and/or gels for Fig. 6.
Source Data Fig. 6
Statistical source data for Fig. 6.
Source Data Fig. 7
Statistical source data for Fig. 7.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, S., Liu, X., Zhang, Z. et al. JMJD2 regulates enhancer–promoter interactions via biomolecular condensate formation. Nat Genet (2025). https://doi.org/10.1038/s41588-025-02415-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41588-025-02415-8