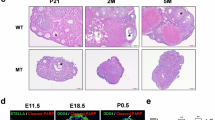
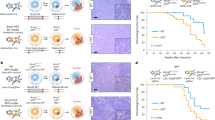
Abstract
Since the discovery of the BRCA1 and BRCA2 (hereafter referred to as BRCA1/2) hereditary breast and ovarian cancer genes three decades ago, genetically engineered and patient-derived mouse models have been instrumental in advancing our understanding of BRCA1/2 biology, particularly their roles in normal development, tumor suppression and therapy response. Brca1/2-mutant mouse models and derivative cell lines have facilitated in vivo dissection of BRCA1/2 functions and identification of the cellular origin and (epi)genetic drivers of BRCA1/2-associated cancer. Genetically engineered and patient-derived mouse tumor models have also been instrumental in developing new (combination) therapies for patients with BRCA1/2-mutated cancers and to study mechanisms of therapy resistance. In this Perspective, we highlight the crucial insights into the complex biology of BRCA1/2 these models have afforded and emphasize those aspects that remain to be elucidated. We also propose next-generation mouse models to further advance our understanding of BRCA1/2 and improve the quality of life of mutation carriers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Capecchi, M. R. Altering the genome by homologous recombination. Science 244, 1288–1292 (1989).
Gowen, L. C., Johnson, B. L., Latour, A. M., Sulik, K. K. & Koller, B. H. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 12, 191–194 (1996).
Hakem, R. et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85, 1009–1023 (1996).
Liu, C. Y., Flesken-Nikitin, A., Li, S., Zeng, Y. & Lee, W. H. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 10, 1835–1843 (1996).
Ludwig, T., Chapman, D. L., Papaioannou, V. E. & Efstratiadis, A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 11, 1226–1241 (1997).
Sharan, S. K. et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386, 804–810 (1997).
Suzuki, A. et al. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 11, 1242–1252 (1997).
Shen, S. X. et al. A targeted disruption of the murine Brca1 gene causes γ-irradiation hypersensitivity and genetic instability. Oncogene 17, 3115–3124 (1998).
Xu, X. et al. Centrosome amplification and a defective G2–M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3, 389–395 (1999).
Connor, F. et al. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat. Genet. 17, 423–430 (1997).
Patel, K. J. et al. Involvement of Brca2 in DNA repair. Mol. Cell 1, 347–357 (1998).
Tutt, A. et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 20, 4704–4716 (2001).
Lomonosov, M., Anand, S., Sangrithi, M., Davies, R. & Venkitaraman, A. R. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 17, 3017–3022 (2003).
Schlacher, K. et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542 (2011).
Schlacher, K., Wu, H. & Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51–BRCA1/2. Cancer Cell 22, 106–116 (2012).
Hatchi, E. et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 57, 636–647 (2015).
Shivji, M. K. K., Renaudin, X., Williams, C. H. & Venkitaraman, A. R. BRCA2 regulates transcription elongation by RNA polymerase II to prevent R-loop accumulation. Cell Rep. 22, 1031–1039 (2018).
Zhang, X. et al. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat. Commun. 8, 15908 (2017).
Patel, P. S. et al. RNF168 regulates R-loop resolution and genomic stability in BRCA1/2-deficient tumors. J. Clin. Invest. 131, e140105 (2021).
Xu, X., Aprelikova, O., Moens, P., Deng, C.-X. & Furth, P. A. Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development 130, 2001–2012 (2003).
Broering, T. J. et al. BRCA1 establishes DNA damage signaling and pericentric heterochromatin of the X chromosome in male meiosis. J. Cell Biol. 205, 663–675 (2014).
Sharan, S. K. et al. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131, 131–142 (2004).
Hakem, R., de la Pompa, J. L., Elia, A., Potter, J. & Mak, T. W. Partial rescue of Brca15–6 early embryonic lethality by p53 or p21 null mutation. Nat. Genet. 16, 298–302 (1997).
Xu, X. et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat. Genet. 28, 266–271 (2001).
Cao, L. et al. ATM–Chk2–p53 activation prevents tumorigenesis at an expense of organ homeostasis upon Brca1 deficiency. EMBO J. 25, 2167–2177 (2006).
Cao, L. et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell 35, 534–541 (2009).
Bouwman, P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695 (2010).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Petrucelli, N., Daly, M. B. & Pal, T. In GeneReviews (eds Adam, M. P. et al.) (University of Washington, 1998–2025).
Ludwig, T., Fisher, P., Ganesan, S. & Efstratiadis, A. Tumorigenesis in mice carrying a truncating Brca1 mutation. Genes Dev. 15, 1188–1193 (2001).
Bachelier, R. et al. Normal lymphocyte development and thymic lymphoma formation in Brca1 exon-11-deficient mice. Oncogene 22, 528–537 (2003).
Kim, S. S. et al. Hyperplasia and spontaneous tumor development in the gynecologic system in mice lacking the BRCA1-Δ11 isoform. Mol. Cell. Biol. 26, 6983–6992 (2006).
Friedman, L. S. et al. Thymic lymphomas in mice with a truncating mutation in Brca2. Cancer Res. 58, 1338–1343 (1998).
McAllister, K. A. et al. Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res. 62, 990–994 (2002).
Xu, X. et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat. Genet. 22, 37–43 (1999).
Liu, X. et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl Acad. Sci. USA 104, 12111–12116 (2007).
McCarthy, A. et al. A mouse model of basal-like breast carcinoma with metaplastic elements. J. Pathol. 211, 389–398 (2007).
Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001).
Ludwig, T., Fisher, P., Murty, V. & Efstratiadis, A. Development of mammary adenocarcinomas by tissue-specific knockout of Brca2 in mice. Oncogene 20, 3937–3948 (2001).
Szabova, L. et al. Perturbation of Rb, p53, and Brca1 or Brca2 cooperate in inducing metastatic serous epithelial ovarian cancer. Cancer Res. 72, 4141–4153 (2012).
Perets, R. et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell 24, 751–765 (2013).
Levanon, K., Crum, C. & Drapkin, R. New insights into the pathogenesis of serous ovarian cancer and its clinical impact. J. Clin. Oncol. 26, 5284–5293 (2008).
Iyer, S. et al. Genetically defined syngeneic mouse models of ovarian cancer as tools for the discovery of combination immunotherapy. Cancer Discov. 11, 384–407 (2021).
Skoulidis, F. et al. Germline Brca2 heterozygosity promotes KrasG12D-driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell 18, 499–509 (2010).
Rowley, M. et al. Inactivation of Brca2 promotes Trp53-associated but inhibits KrasG12D-dependent pancreatic cancer development in mice. Gastroenterology 140, 1303–1313 (2011).
Shakya, R. et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 334, 525–528 (2011).
Drost, R. et al. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell 20, 797–809 (2011).
Drost, R. et al. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J. Clin. Invest. 126, 2903–2918 (2016).
Chen, J. et al. 53BP1 loss rescues embryonic lethality but not genomic instability of BRCA1 total knockout mice. Cell Death Differ. 27, 2552–2567 (2020).
Nacson, J. et al. BRCA1 mutational complementation induces synthetic viability. Mol. Cell 78, 951–959 (2020).
Park, D. et al. Ablation of the Brca1–Palb2 interaction phenocopies Fanconi anemia in mice. Cancer Res. 80, 4172–4184 (2020).
Pulver, E. M. et al. A BRCA1 coiled-coil domain variant disrupting PALB2 interaction promotes the development of mammary tumors and confers a targetable defect in homologous recombination repair. Cancer Res. 81, 6171–6182 (2021).
Krais, J. J. et al. RNF168-mediated localization of BARD1 recruits the BRCA1–PALB2 complex to DNA damage. Nat. Commun. 12, 5016 (2021).
Hartford, S. A. et al. Interaction with PALB2 is essential for maintenance of genomic integrity by BRCA2. PLoS Genet. 12, e1006236 (2016).
Lim, P. X., Zaman, M., Feng, W. & Jasin, M. BRCA2 promotes genomic integrity and therapy resistance primarily through its role in homology-directed repair. Mol. Cell 84, 447–462 (2024).
Atanassov, B. S., Barrett, J. C. & Davis, B. J. Homozygous germ line mutation in exon 27 of murine Brca2 disrupts the Fancd2–Brca2 pathway in the homologous recombination-mediated DNA interstrand cross-links’ repair but does not affect meiosis. Genes Chromosomes Cancer 44, 429–437 (2005).
Pathania, S. et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat. Commun. 5, 5496 (2014).
Konishi, H. et al. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proc. Natl Acad. Sci. USA 108, 17773–17778 (2011).
Sedic, M. et al. Haploinsufficiency for BRCA1 leads to cell-type-specific genomic instability and premature senescence. Nat. Commun. 6, 7505 (2015).
Zong, D. et al. BRCA1 haploinsufficiency is masked by RNF168-mediated chromatin ubiquitylation. Mol. Cell 73, 1267–1281 (2019).
Li, C. M. et al. Brca1 haploinsufficiency promotes early tumor onset and epigenetic alterations in a mouse model of hereditary breast cancer. Nat. Genet. 56, 2763–2775 (2024).
Kumar, P. et al. Cooperativity of Rb, Brca1, and p53 in malignant breast cancer evolution. PLoS Genet. 8, e1003027 (2012).
Annunziato, S. et al. Comparative oncogenomics identifies combinations of driver genes and drug targets in BRCA1-mutated breast cancer. Nat. Commun. 10, 397 (2019).
Liu, H. et al. Identifying and targeting sporadic oncogenic genetic aberrations in mouse models of triple-negative breast cancer. Cancer Discov. 8, 354–369 (2018).
Menghi, F. et al. The tandem duplicator phenotype is a prevalent genome-wide cancer configuration driven by distinct gene mutations. Cancer Cell 34, 197–210 (2018).
Molyneux, G. et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7, 403–417 (2010).
Sau, A. et al. Persistent activation of NF-κB in BRCA1-deficient mammary progenitors drives aberrant proliferation and accumulation of DNA damage. Cell Stem Cell 19, 52–65 (2016).
Bach, K. et al. Time-resolved single-cell analysis of Brca1 associated mammary tumourigenesis reveals aberrant differentiation of luminal progenitors. Nat. Commun. 12, 1502 (2021).
Ciwinska, M. et al. Mechanisms that clear mutations drive field cancerization in mammary tissue. Nature 633, 198–206 (2024).
Rottenberg, S. et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc. Natl Acad. Sci. USA 104, 12117–12122 (2007).
Zander, S. A. et al. Sensitivity and acquired resistance of BRCA1;p53-deficient mouse mammary tumors to the topoisomerase I inhibitor topotecan. Cancer Res. 70, 1700–1710 (2010).
Pajic, M. et al. Tumor-initiating cells are not enriched in cisplatin-surviving BRCA1;p53-deficient mammary tumor cells in vivo. Cell Cycle 9, 3780–3791 (2010).
Pajic, M. et al. Selected alkylating agents can overcome drug tolerance of G0-like tumor cells and eradicate BRCA1-deficient mammary tumors in mice. Clin. Cancer Res. 23, 7020–7033 (2017).
Evers, B. et al. A high-throughput pharmaceutical screen identifies compounds with specific toxicity against BRCA2-deficient tumors. Clin. Cancer Res. 16, 99–108 (2010).
Planells-Cases, R. et al. Subunit composition of VRAC channels determines substrate specificity and cellular resistance to Pt-based anti-cancer drugs. EMBO J. 34, 2993–3008 (2015).
Widmer, C. A. et al. Loss of the volume-regulated anion channel components LRRC8A and LRRC8D limits platinum drug efficacy. Cancer Res. Commun. 2, 1266–1281 (2022).
Rottenberg, S. et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl Acad. Sci. USA 105, 17079–17084 (2008).
Hay, T. et al. Poly(ADP-ribose) polymerase-1 inhibitor treatment regresses autochthonous Brca2/p53-mutant mammary tumors in vivo and delays tumor relapse in combination with carboplatin. Cancer Res. 69, 3850–3855 (2009).
Jaspers, J. E. et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 3, 68–81 (2013).
Xu, G. et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544 (2015).
Barazas, M. et al. The CST complex mediates end protection at double-strand breaks and promotes PARP inhibitor sensitivity in BRCA1-deficient cells. Cell Rep. 23, 2107–2118 (2018).
Ter Brugge, P. et al. Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. J. Natl Cancer Inst. 108, djw148 (2016).
Gogola, E. et al. Selective Loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell 33, 1078–1093 (2018).
Paes Dias, M. et al. Loss of nuclear DNA ligase III reverts PARP inhibitor resistance in BRCA1/53BP1 double-deficient cells by exposing ssDNA gaps. Mol. Cell 81, 4692–4708 (2021).
Barazas, M. et al. Radiosensitivity is an acquired vulnerability of PARPi-resistant BRCA1-deficient tumors. Cancer Res. 79, 452–460 (2019).
Juvekar, A. et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2, 1048–1063 (2012).
Ding, L. et al. PARP inhibition elicits STING-dependent antitumor immunity in Brca1-deficient ovarian cancer. Cell Rep. 25, 2972–2980 (2018).
Pantelidou, C. et al. PARP inhibitor efficacy depends on CD8+ T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 9, 722–737 (2019).
Wang, Q. et al. STING agonism reprograms tumor-associated macrophages and overcomes resistance to PARP inhibition in BRCA1-deficient models of breast cancer. Nat. Commun. 13, 3022 (2022).
Zatreanu, D. et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun. 12, 3636 (2021).
Zhou, J. et al. A first-in-class polymerase θ inhibitor selectively targets homologous-recombination-deficient tumors. Nat. Cancer 2, 598–610 (2021).
Mateos-Gomez, P. A. et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015).
Ceccaldi, R. et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 (2015).
Krais, J. J. et al. Genetic separation of Brca1 functions reveal mutation-dependent Polθ vulnerabilities. Nat. Commun. 14, 7714 (2023).
DuPage, M. & Jacks, T. Genetically engineered mouse models of cancer reveal new insights about the antitumor immune response. Curr. Opin. Immunol. 25, 192–199 (2013).
Nolan, E. et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci. Transl. Med. 9, eaal4922 (2017).
Rebbeck, T. R. et al. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N. Engl. J. Med. 346, 1616–1622 (2002).
Poole, A. J. et al. Prevention of Brca1-mediated mammary tumorigenesis in mice by a progesterone antagonist. Science 314, 1467–1470 (2006).
Nolan, E. et al. RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat. Med. 22, 933–939 (2016).
Sigl, V. et al. RANKL/RANK control Brca1 mutation-driven mammary tumors. Cell Res. 26, 761–774 (2016).
van de Ven, M. et al. BRCA1-associated mammary tumorigenesis is dependent on estrogen rather than progesterone signaling. J. Pathol. 246, 41–53 (2018).
Gorrini, C. et al. Estrogen controls the survival of BRCA1-deficient cells via a PI3K–NRF2-regulated pathway. Proc. Natl Acad. Sci. USA 111, 4472–4477 (2014).
Vinayak, S. & Ford, J. M. PARP inhibitors for the treatment and prevention of breast cancer. Curr. Breast Cancer Rep. 2, 190–197 (2010).
To, C. et al. The PARP inhibitors, veliparib and olaparib, are effective chemopreventive agents for delaying mammary tumor development in BRCA1-deficient mice. Cancer Prev. Res. 7, 698–707 (2014).
van de Ven, M. et al. Prophylactic window therapy with the clinical poly(ADP-ribose) polymerase inhibitor olaparib delays BRCA1-deficient mammary tumour formation in mice. J. Pathol. 241, 511–521 (2017).
Zeng, J. et al. A genetic mosaic mouse model illuminates the pre-malignant progression of basal-like breast cancer. Dis. Model. Mech. 16, dmm050219 (2023).
Singh, A. K. & Yu, X. Tissue-specific carcinogens as soil to seed BRCA1/2-mutant hereditary cancers. Trends Cancer 6, 559–568 (2020).
Kass, E. M., Lim, P. X., Helgadottir, H. R., Moynahan, M. E. & Jasin, M. Robust homology-directed repair within mouse mammary tissue is not specifically affected by Brca2 mutation. Nat. Commun. 7, 13241 (2016).
Uijttewaal, E. C. H. et al. CRISPR-StAR enables high-resolution genetic screening in complex in vivo models. Nat. Biotechnol. 43, 1848–1860 (2024).
Yamashita, M. et al. Cell-type specific, inducible and acute degradation of targeted protein in mice by two degron systems. Nat. Commun. 15, 10129 (2024).
Pettitt, S. J. et al. Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discov. 10, 1475–1488 (2020).
Billing, D. et al. The BRCT domains of the BRCA1 and BARD1 tumor suppressors differentially regulate homology-directed repair and stalled fork protection. Mol. Cell 72, 127–139 (2018).
Stratton, M. R. Pathology of familial breast cancer: differences between breast cancers in carriers of BRCA1 or BRCA2 mutations and sporadic cases. Lancet 349, 1505–1510 (1997).
Lakhani, S. R. et al. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J. Natl Cancer Inst. 90, 1138–1145 (1998).
Bhin, J. et al. Multi-omics analysis reveals distinct non-reversion mechanisms of PARPi resistance in BRCA1- versus BRCA2-deficient mammary tumors. Cell Rep. 42, 112538 (2023).
Holstege, H. et al. Cross-species comparison of aCGH data from mouse and human BRCA1- and BRCA2-mutated breast cancers. BMC Cancer 10, 455 (2010).
Acknowledgements
We apologize to those authors whose work could not be cited due to space constraints. Research at the Netherlands Cancer Institute is supported by international grants from the Dutch Cancer Society (KWF) and the Dutch Ministry of Health, Welfare and Sport. Research in the Jonkers laboratory is funded by the Oncode Institute, the Dutch Cancer Society (grants 14516 and 14949) and the Swiss National Science Foundation (grant 320030M_219453).
Author information
Authors and Affiliations
Contributions
All authors contributed to writing the Perspective and reviewing and editing the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
J.J. reports funding from Artios Pharma outside of the submitted work. J.-S.D. has no competing interests to declare.
Peer review
Peer review information
Nature Genetics thanks Hui Zong and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Supplementary References
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Darnold, JS., Jonkers, J. Insights from three decades of BRCA1/2 modeling in mice. Nat Genet (2026). https://doi.org/10.1038/s41588-025-02448-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41588-025-02448-z