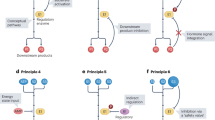
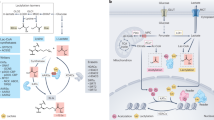
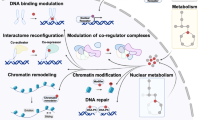
Abstract
Intricate coupling between metabolism and protein post-translational modifications (PTMs) has emerged as a fundamental aspect of cellular regulation. Recent studies demonstrate that protein modifications can originate from diverse metabolites, and that their regulation is closely tied to the cellular metabolic state. Here we explore recently uncovered PTMs, including the concept of ‘modification of a modification’, as well as associated feedback and feedforward regulatory mechanisms, in which modified proteins impact not only related metabolic pathways but also other signaling cascades affecting physiology and diseases. The recently uncovered role of nucleus-localized metabolic enzymes for histone modifications additionally highlights the importance of cell-compartment-specific metabolic states. We further comment on the utility of untargeted metabolomics and proteomics for previously unrecognized PTMs and associated metabolic patterns. Together, these advances have uncovered a dynamic interplay between metabolism and PTMs, offering new perspectives for understanding metabolic regulation and developing targeted therapeutic strategies.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
No data were generated for this manuscript.
References
Keenan, E. K., Zachman, D. K. & Hirschey, M. D. Discovering the landscape of protein modifications. Mol. Cell 81, 1868–1878 (2021).
Gyorgy, B., Toth, E., Tarcsa, E., Falus, A. & Buzas, E. I. Citrullination: a posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 38, 1662–1677 (2006).
Shang, S., Liu, J. & Hua, F. Protein acylation: mechanisms, biological functions and therapeutic targets. Signal Transduct. Target Ther. 7, 396 (2022).
Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019).
Zhu, Z. et al. Identification of lysine isobutyrylation as a new histone modification mark. Nucleic Acids Res. 49, 177–189 (2021).
Gao, Y. et al. Identification of histone lysine acetoacetylation as a dynamic post-translational modification regulated by HBO1. Adv. Sci. 10, e2300032 (2023).
Liu, D. et al. Discovery of itaconate-mediated lysine acylation. J. Am. Chem. Soc. 145, 12673–12681 (2023).
Delaney, K. et al. Histone lysine methacrylation is a dynamic post-translational modification regulated by HAT1 and SIRT2. Cell Discov. 7, 122 (2021).
Huang, H. et al. Lysine benzoylation is a histone mark regulated by SIRT2. Nat. Commun. 9, 3374 (2018).
Jiang, Y. H. et al. Isonicotinylation is a histone mark induced by the anti-tuberculosis first-line drug isoniazid. Nat. Commun. 12, 5548 (2021).
Moellering, R. E. & Cravatt, B. F. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science 341, 549–553 (2013).
Hu, S. H. et al. Amino acids downregulate SIRT4 to detoxify ammonia through the urea cycle. Nat. Metab. 5, 626–641 (2023).
Keenan, E. K., Bareja, A., Lam, Y., Grimsrud, P. A. & Hirschey, M. D. Cysteine S-acetylation is a post-translational modification involved in metabolic regulation. Preprint at bioRxiv https://doi.org/10.1101/2024.05.21.595030 (2024).
Zheng, Q. et al. Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat. Commun. 10, 1289 (2019).
Zhang, H. & Forman, H. J. Signaling by 4-hydroxy-2-nonenal: exposure protocols, target selectivity and degradation. Arch. Biochem. Biophys. 617, 145–154 (2017).
Hu, S. H. et al. Methylene-bridge tryptophan fatty acylation regulates PI3K-AKT signaling and glucose uptake. Cell Rep 38, 110509 (2022). This paper describes a chemically unusual PTM that is derived from polyunsaturated fatty acids. The modification was shown to occur at important kinases PDK1 and AKT2, regulating cell signaling.
Linthwaite, V. L. et al. Ubiquitin is a carbon dioxide-binding protein. Sci. Adv. 7, eabi5507 (2021).
Pham, V. N. et al. Formaldehyde regulates S-adenosylmethionine biosynthesis and one-carbon metabolism. Science 382, eabp9201 (2023).
Seim, G. L. et al. Nitric oxide-driven modifications of lipoic arm inhibit alpha-ketoacid dehydrogenases. Nat. Chem. Biol. 19, 265–274 (2023). This study shows that the production of reactive nitrogen species and cellular metabolic changes are linked via covalent modification of a metabolic enzyme-attached lipoyl group, as a representative example of modification-of-a-modification.
Arp, N. L., Seim, G. L., Votava, J. A., Josephson, J. & Fan, J. Reactive nitrogen species inhibit branched chain α-ketoacid dehydrogenase complex and impact muscle cell metabolism. J. Biol. Chem. 299, 105333 (2023).
Minogue, E. et al. Glutarate regulates T cell metabolism and anti-tumour immunity. Nat. Metab. 5, 1747–1764 (2023).
Lu-Culligan, W. J. et al. Acetyl-methyllysine marks chromatin at active transcription start sites. Nature 622, 173–179 (2023).
Ma, B. B. et al. Post-crotonylation oxidation by a monooxygenase promotes acetyl-CoA synthetase degradation in Streptomyces roseosporus. Commun. Biol. 6, 1243 (2023).
Carrico, C., Meyer, J. G., He, W., Gibson, B. W. & Verdin, E. The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications. Cell Metab. 27, 497–512 (2018).
Ma, W. H. et al. OXCT1 functions as a succinyltransferase, contributing to hepatocellular carcinoma via succinylating LACTB. Mol. Cell 84, 538–551 (2024).
Wagner, G. R. et al. A class of reactive acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metab. 25, 823–837.e828 (2017).
Simic, Z., Weiwad, M., Schierhorn, A., Steegborn, C. & Schutkowski, M. The ɛ-amino group of protein lysine residues is highly susceptible to nonenzymatic acylation by several physiological acyl-CoA thioesters. ChemBioChem 16, 2337–2347 (2015).
Kulkarni, R. A. et al. Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chem. Biol. 24, 231–242 (2017).
Bhatt, D. P. et al. Deglutarylation of glutaryl-CoA dehydrogenase by deacylating enzyme SIRT5 promotes lysine oxidation in mice. J. Biol. Chem. 298, 101723 (2022). This study provides insight into specific acylation in an acyl-CoA-binding protein. It also demonstrates coupling between protein glutarylation and metabolism of the free amino acid lysine.
Lu, X. et al. OGDH mediates the inhibition of SIRT5 on cell proliferation and migration of gastric cancer. Exp. Cell. Res. 382, 111483 (2019).
Cao, H. Y. et al. Malonylation of acetyl-CoA carboxylase 1 promotes hepatic steatosis and is attenuated by ketogenic diet in NAFLD. Cell Rep 42, 112319 (2023).
Carrico, C. et al. Coenzyme A binding sites induce proximal acylation across protein families. Sci. Rep. 13, 5029 (2023).
Sjoblom, N. M., Kelsey, M. M. G. & Scheck, R. A. A systematic study of selective protein glycation. Angew. Chem. Int. Ed. 57, 16077–16082 (2018).
Tzeng, S. C. & Maier, C. S. Label-free proteomics assisted by affinity enrichment for elucidating the chemical reactivity of the liver mitochondrial proteome toward adduction by the lipid electrophile 4-hydroxy-2-nonenal (HNE). Front. Chem. 4, 2 (2016).
McEwen, J. M., Fraser, S., Guir, A. L. S., Dave, J. & Scheck, R. A. Synergistic sequence contributions bias glycation outcomes. Nat. Commun. 12, 3316 (2021).
Szapacs, M. E., Riggins, J. N., Zimmerman, L. J. & Liebler, D. C. Covalent adduction of human serum albumin by 4-hydroxy-2-nonenal: kinetic analysis of competing alkylation reactions. Biochemistry 45, 10521–10528 (2006).
Pedley, A. M., Pareek, V. & Benkovic, S. J. The purinosome: a case study for a mammalian metabolon. Annu. Rev. Biochem. 91, 89–106 (2022).
Martell, E. et al. Metabolism-based targeting of MYC via MPC-SOD2 axis-mediated oxidation promotes cellular differentiation in group 3 medulloblastoma. Nat. Commun. 14, 2502 (2023).
Park, K. C. et al. Disrupted propionate metabolism evokes transcriptional changes in the heart by increasing histone acetylation and propionylation. Nat. Cardiovasc. Res. 2, 1221–1245 (2023).
Lagerwaard, B. et al. Increased protein propionylation contributes to mitochondrial dysfunction in liver cells and fibroblasts, but not in myotubes. J. Inherit. Metab. Dis. 44, 438–449 (2021).
Head, P. E. et al. Aberrant methylmalonylation underlies methylmalonic acidemia and is attenuated by an engineered sirtuin. Sci. Transl. Med. 14, eabn4772 (2022).
Zhang, B. et al. Acylspermidines are conserved mitochondrial sirtuin-dependent metabolites. Nat. Chem. Biol. 20, 812–822 (2024). This paper describes a family of metabolites that are downstream of protein lysine deacylation activity by mitochondrial sirtuins, demonstrating how untargeted metabolomics can reveal the substrate of an eraser enzyme.
Palmieri, E. M. et al. Pyruvate dehydrogenase operates as an intramolecular nitroxyl generator during macrophage metabolic reprogramming. Nat. Commun. 14, 5114 (2023).
Hibbs, J. B. Jr., Taintor, R. R., Vavrin, Z. & Rachlin, E. M. Nitric oxide: a cytotoxic activated macrophage effector molecule. Biochem. Biophys. Res. Commun. 157, 87–94 (1988).
Zhang, X. et al. Homocysteine inhibits pro-insulin receptor cleavage and causes insulin resistance via protein cysteine-homocysteinylation. Cell Rep. 37, 109821 (2021).
Aboushousha, R. et al. Glutaredoxin attenuates glutathione levels via deglutathionylation of Otub1 and subsequent destabilization of system xC−. Sci. Adv. 9, eadi5192 (2023).
Yi, W. et al. Protein S-nitrosylation regulates proteostasis and viability of hematopoietic stem cell during regeneration. Cell Rep. 34, 108922 (2021).
Francois, C. M., Pihl, T., Dunoyer de Segonzac, M., Herault, C. & Hudry, B. Metabolic regulation of proteome stability via N-terminal acetylation controls male germline stem cell differentiation and reproduction. Nat. Commun. 14, 6737 (2023).
Varland, S. et al. N-terminal acetylation levels are maintained during acetyl-CoA deficiency in Saccharomyces cerevisiae. Mol. Cell Proteom. 17, 2309–2323 (2018).
Bishop, T. R. et al. Acetyl-CoA biosynthesis drives resistance to histone acetyltransferase inhibition. Nat. Chem. Biol. 19, 1215–1222 (2023).
Pan, R. Y. et al. Positive feedback regulation of microglial glucose metabolism by histone H4 lysine 12 lactylation in Alzheimer’s disease. Cell Metab. 34, 634–648.e636 (2022).
Talwadekar, M. et al. Metabolic transitions regulate global protein fatty acylation. J. Biol. Chem. 300, 105563 (2024).
Nuskova, H. et al. Stearic acid blunts growth-factor signaling via oleoylation of GNAI proteins. Nat. Commun. 12, 4590 (2021).
Zhang, B. et al. Amino acid and protein specificity of protein fatty acylation in C. elegans. Proc. Natl Acad. Sci. USA 121, e2307515121 (2024). This study demonstrates that Lys, Ser and Cys residues are preferentially modified with fatty acyls derived from distinct branches of lipid metabolism.
Yu, Y. et al. An untargeted approach for revealing electrophilic metabolites. ACS Chem. Biol. 15, 3030–3037 (2020).
Tang, H. Y., Cui, M. X. & Han, M. Fatty acids impact sarcomere integrity through myristoylation and ER homeostasis. Cell Rep 36, 109539 (2021).
Kallemeijn, W. W. et al. Proteome-wide analysis of protein lipidation using chemical probes: in-gel fluorescence visualization, identification and quantification of N-myristoylation, N- and S-acylation, O-cholesterylation, S-farnesylation and S-geranylgeranylation. Nat. Protoc. 16, 5083–5122 (2021).
Liu, Y. et al. MAVS Cys508 palmitoylation promotes its aggregation on the mitochondrial outer membrane and antiviral innate immunity. Proc. Natl Acad. Sci. USA 121, e2403392121 (2024).
Karthigeyan, K. P. et al. A bioorthogonal chemical reporter for fatty acid synthase-dependent protein acylation. J. Biol. Chem. 297, 101272 (2021).
Puleston, D. J. et al. Polyamine metabolism is a central determinant of helper T cell lineage fidelity. Cell 184, 4186–4202.e4120 (2021). This paper identifies the coupling between polyamine synthesis and eIF5A hypusination when CD4+ T cells differentiate, which is required for T helper cell lineage fidelity.
Zhou, J. et al. Spermidine-mediated hypusination of translation factor EIF5A improves mitochondrial fatty acid oxidation and prevents non-alcoholic steatohepatitis progression. Nat. Commun. 13, 5202 (2022).
Li, H. et al. YAP/TAZ drives cell proliferation and tumour growth via a polyamine-eIF5A hypusination-LSD1 axis. Nat. Cell Biol. 24, 373–383 (2022).
Choi, U. Y. et al. Herpesvirus-induced spermidine synthesis and eIF5A hypusination for viral episomal maintenance. Cell Rep. 40, 111234 (2022).
Mahalingam, S. S. et al. Polyamine metabolism impacts T cell dysfunction in the oral mucosa of people living with HIV. Nat. Commun. 14, 399 (2023).
Sun, R. C. et al. Brain glycogen serves as a critical glucosamine cache required for protein glycosylation. Cell Metab. 33, 1404–1417 (2021).
Liu, X. et al. Hexosamine biosynthetic pathway and O-GlcNAc-processing enzymes regulate daily rhythms in protein O-GlcNAcylation. Nat. Commun. 12, 4173 (2021).
Shi, Q. et al. Increased glucose metabolism in TAMs fuels O-GlcNAcylation of lysosomal Cathepsin B to promote cancer metastasis and chemoresistance. Cancer Cell 40, 1207–1222 e1210 (2022).
Petrus, P. et al. Glutamine links obesity to inflammation in human white adipose tissue. Cell Metab. 31, 375–390 e311 (2020).
Liu, X., Cai, Y. D. & Chiu, J. C. Regulation of protein O-GlcNAcylation by circadian, metabolic and cellular signals. J. Biol. Chem. 300, 105616 (2024).
Wang, X. et al. A GAPDH serotonylation system couples CD8+ T cell glycolytic metabolism to antitumor immunity. Mol. Cell 84, 760–775 e767 (2024).
Mordhorst, A. et al. Phenylalanine hydroxylase contributes to serotonin synthesis in mice. FASEB J. 35, e21648 (2021).
Yu, J. et al. Parallel pathways for serotonin biosynthesis and metabolism in C. elegans. Nat. Chem. Biol. 19, 141–150 (2023).
Li, H. et al. β-hydroxybutyrate reduces reinstatement of cocaine conditioned place preference through hippocampal CaMKII-α β-hydroxybutyrylation. Cell Rep. 41, 111724 (2022).
Lund, P. J. et al. Stable isotope tracing in vivo reveals a metabolic bridge linking the microbiota to host histone acetylation. Cell Rep. 41, 111809 (2022).
Zhou, Q. et al. Phenylalanine impairs insulin signaling and inhibits glucose uptake through modification of IRβ. Nat. Commun. 13, 4291 (2022).
He, X. D. et al. Sensing and transmitting intracellular amino acid signals through reversible lysine aminoacylations. Cell Metab. 27, 151–166 (2018).
Zong, Z. et al. Alanyl-tRNA synthetase, AARS1, is a lactate sensor and lactyltransferase that lactylates p53 and contributes to tumorigenesis. Cell 187, 2375–2392 e2333 (2024). This paper shows that one of the ARSs can act as a lactyltransferase, which potentially effects a broad range of lactylation-regulated processes. Together with He et al. (2018), these findings provide notable examples of the coupling of tumor cell metabolism to protein modifications and demonstrate that the roles of ARSs as PTM writers extend beyond previously reported lysine amino acylation.
Howie, J. et al. Glutathione-dependent depalmitoylation of phospholemman by peroxiredoxin 6. Cell Rep. 43, 113679 (2024).
Martin-Gallausiaux, C., Marinelli, L., Blottiere, H. M., Larraufie, P. & Lapaque, N. SCFA: mechanisms and functional importance in the gut. Proc. Nutr. Soc. 80, 37–49 (2021).
Thomas, S. P. & Denu, J. M. Short-chain fatty acids activate acetyltransferase p300. eLife 10, e72171 (2021).
Nitsch, S., Zorro Shahidian, L. & Schneider, R. Histone acylations and chromatin dynamics: concepts, challenges and links to metabolism. EMBO Rep. 22, e52774 (2021).
Qin, F. et al. Linking chromatin acylation mark-defined proteome and genome in living cells. Cell 186, 1066–1085 e1036 (2023).
Diehl, K. L. & Muir, T. W. Chromatin as a key consumer in the metabolite economy. Nat. Chem. Biol. 16, 620–629 (2020).
Trefely, S., Lovell, C. D., Snyder, N. W. & Wellen, K. E. Compartmentalised acyl-CoA metabolism and roles in chromatin regulation. Mol. Metab. 38, 100941 (2020).
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009).
Nagaraj, R. et al. Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell 168, 210–223 e211 (2017).
Li, W. et al. Nuclear localization of mitochondrial TCA cycle enzymes modulates pluripotency via histone acetylation. Nat. Commun. 13, 7414 (2022).
Bulusu, V. et al. Acetate recapturing by nuclear acetyl-CoA synthetase 2 prevents loss of histone acetylation during oxygen and serum limitation. Cell Rep. 18, 647–658 (2017).
Mendoza, M. et al. Enzymatic transfer of acetate on histones from lysine reservoir sites to lysine activating sites. Sci. Adv. 8, eabj5688 (2022).
Shvedunova, M. & Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23, 329–349 (2022).
Singh, S., Senapati, P. & Kundu, T. K. Metabolic regulation of lysine acetylation: implications in cancer. Subcell. Biochem. 100, 393–426 (2022).
Kafkia, E. et al. Operation of a TCA cycle subnetwork in the mammalian nucleus. Sci. Adv. 8, eabq5206 (2022). This paper describes the presence of a part of the TCA cycle in the nucleus and demonstrates its contribution to nuclear protein succinylation. The finding was based on metabolomic analysis of the nucleus.
Liu, X. J. et al. The existence of a nonclassical TCA cycle in the nucleus that wires the metabolic-epigenetic circuitry. Signal Transduct. Target Ther 6, 375 (2021).
Wang, Y. et al. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 (2017).
Yuan, H. R. et al. Lysine catabolism reprograms tumour immunity through histone crotonylation. Nature 617, 818–826 (2023). This paper describes the presence of a key lysine catabolism enzyme in the nucleus and shows that the product of the enzyme (that is, crotonyl-CoA) contributes to histone modification. This hypothesis was developed from the observation of lysine uptake changes in the glioblastoma stem cells revealed by metabolomics.
Anmangandla, A., Ren, Y., Fu, Q., Zhang, S. & Lin, H. The acyl-CoA specificity of human lysine acetyltransferase KAT2A. Biochemistry 61, 1874–1882 (2022).
Russo, M. et al. Acetyl-CoA production by mediator-bound 2-ketoacid dehydrogenases boosts de novo histone acetylation and is regulated by nitric oxide. Mol. Cell 84, 967–980 e910 (2024).
Zervopoulos, S. D. et al. MFN2-driven mitochondria-to-nucleus tethering allows a non-canonical nuclear entry pathway of the mitochondrial pyruvate dehydrogenase complex. Mol. Cell 82, 1066–1077 e1067 (2022).
Fransen, M., Lismont, C. & Walton, P. The peroxisome–mitochondria connection: how and why?. Int. J. Mol. Sci. 18, 1126 (2017).
Yang, Y. et al. Altered succinylation of mitochondrial proteins, APP and tau in Alzheimer’s disease. Nat. Commun. 13, 159 (2022).
Mills, E. L. et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018).
Huang, Z. W. et al. STAT5 promotes PD-L1 expression by facilitating histone lactylation to drive immunosuppression in acute myeloid leukemia. Signal Transduct. Target Ther. 8, 391 (2023).
Gut, P. et al. SUCLA2 mutations cause global protein succinylation contributing to the pathomechanism of a hereditary mitochondrial disease. Nat. Commun. 11, 5927 (2020).
Zhou, P. et al. High dietary fructose promotes hepatocellular carcinoma progression by enhancing O-GlcNAcylation via microbiota-derived acetate. Cell Metab. 35, 1961–1975 e1966 (2023).
Helf, M. J., Fox, B. W., Artyukhin, A. B., Zhang, Y. K. & Schroeder, F. C. Comparative metabolomics with Metaboseek reveals functions of a conserved fat metabolism pathway in C. elegans. Nat. Commun. 13, 782 (2022).
Bern, M., Kil, Y. J. & Becker, C. Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics 13, 13.20.11–13.20.14 (2012).
Zhai, Y. et al. Cysteine carboxyethylation generates neoantigens to induce HLA-restricted autoimmunity. Science 379, eabg2482 (2023). This study uses sequencing-based proteomics data analysis algorithms to look for PTM changes in an untargeted manner (that is, the searching is not limited to a short list of known PTMs, as in conventional workflows).
Geiszler, D. J., Polasky, D. A., Yu, F. & Nesvizhskii, A. I. Detecting diagnostic features in MS/MS spectra of post-translationally modified peptides. Nat. Commun. 14, 4132 (2023).
Gao, J. et al. Identification of 113 new histone marks by CHiMA, a tailored database search strategy. Sci. Adv. 9, eadf1416 (2023).
Prus, G., Hoegl, A., Weinert, B. T. & Choudhary, C. Analysis and interpretation of protein post-translational modification site stoichiometry. Trends Biochem. Sci. 44, 943–960 (2019).
Zhang, R. et al. Regulation of urea cycle by reversible high-stoichiometry lysine succinylation. Nat. Metab. 6, 550–566 (2024).
Trefely, S. et al. Quantitative subcellular acyl-CoA analysis reveals distinct nuclear metabolism and isoleucine-dependent histone propionylation. Mol. Cell 82, 447–462 e446 (2022).
James, A. M., Smith, A. C., Smith, C. L., Robinson, A. J. & Murphy, M. P. Proximal cysteines that enhance lysine acetylation of cytosolic proteins in mice are less conserved in longer-living species. Cell Rep. 24, 1445–1455 (2018).
Acknowledgements
We thank E. N. Garner for comments and proofreading the manuscript. This work was supported, in part, by the National Institutes of Health (R35GM131877).
Author information
Authors and Affiliations
Contributions
B.Z. conceived the idea and outline for this Perspective. B.Z. and F.C.S. wrote the text and prepared figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Shi-Min Zhao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, B., Schroeder, F.C. Mechanisms of metabolism-coupled protein modifications. Nat Chem Biol 21, 819–830 (2025). https://doi.org/10.1038/s41589-024-01805-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41589-024-01805-z