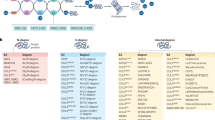
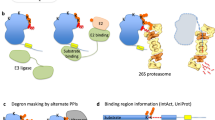
Abstract
Ubiquitin-dependent protein degradation regulates myriad fundamental cellular processes. At its core are degradation signals, or degrons, that initiate substrate engagement and ubiquitination by E3 ubiquitin ligases. Here we highlight how a variety of degradation signals promote substrate–E3 ligase interactions to orchestrate protein turnover with precision. While short linear motifs are frequently identified as degrons, an increasing number of degrons have recently been mapped to high-order protein structures, underscoring the architectural diversity and cryptic nature of degradation signals. Furthermore, nonproteinaceous signals beyond degrons often facilitate the precise control of protein ubiquitination. These additional signals can reside within substrates and E3 ligases or at their interfaces. Finally, we discuss how dysregulation of degrons and degradation signals is linked to human diseases. A deeper mechanistic understanding of degradation signals will guide new therapeutic strategies, whether by restoring defective protein ubiquitination or by harnessing targeted protein degradation.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Müller, L. & Hoppe, T. UPS-dependent strategies of protein quality control degradation. Trends Biochem. Sci. 49, 859–874 (2024).
Hochstrasser, M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30, 405–439 (1996).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Cowan, A. D. & Ciulli, A. Driving E3 ligase substrate specificity for targeted protein degradation: lessons from nature and the laboratory. Annu. Rev. Biochem. 91, 295–319 (2022).
Ravid, T. & Hochstrasser, M. Diversity of degradation signals in the ubiquitin–proteasome system. Nat. Rev. Mol. Cell Biol. 9, 679–690 (2008).
Varshavsky, A. Naming a targeting signal. Cell 64, 13–15 (1991).
Bachmair, A., Finley, D. & Varshavsky, A. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 (1986).
Okoye, C. N., Rowling, P. J. E., Itzhaki, L. S. & Lindon, C. Counting degrons: lessons from multivalent substrates for targeted protein degradation. Front. Physiol. 13, 913063 (2022).
Harris, T. J. & Trader, D. J. Exploration of degrons and their ability to mediate targeted protein degradation. RSC Med. Chem. https://doi.org/10.1039/d4md00787e (2025).
Varshavsky, A. N-degron pathways. Proc. Natl Acad. Sci. USA 121, e2408697121 (2024).
Sherpa, D., Chrustowicz, J. & Schulman, B. A. How the ends signal the end: regulation by E3 ubiquitin ligases recognizing protein termini. Mol. Cell 82, 1424–1438 (2022).
Zheng, N. & Shabek, N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017).
Zhang, Z., Mena, E. L., Timms, R. T., Koren, I. & Elledge, S. J. Degrons: defining the rules of protein degradation. Nat. Rev. Mol. Cell Biol. https://doi.org/10.1038/s41580-025-00870-z (2025).
Padovani, C., Jevtić, P. & Rapé, M. Quality control of protein complex composition. Mol. Cell 82, 1439–1450 (2022).
Rogers, S., Wells, R. & Rechsteiner, M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234, 364–368 (1986).
Skaar, J. R., Pagan, J. K. & Pagano, M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 (2013).
Latres, E., Chiaur, D. S. & Pagano, M. The human F box protein β-Trcp associates with the Cul1/Skp1 complex and regulates the stability of β-catenin. Oncogene 18, 849–854 (1999).
Hart, M. et al. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr. Biol. 9, 207–210 (1999).
Wu, G. et al. Structure of a β-TrCP1–Skp1–β-catenin complex: destruction motif binding and lysine specificity of the SCFβ-TrCP1 ubiquitin ligase. Mol. Cell 11, 1445–1456 (2003).
Kim, T. Y. et al. Substrate trapping proteomics reveals targets of the βTrCP2/FBXW11 ubiquitin ligase. Mol. Cell. Biol. 35, 167–181 (2015).
Ivan, M. et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 (2001).
Lee, J. M. et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell 48, 572–586 (2012).
Taguchi, K. & Yamamoto, M. The KEAP1–NRF2 system in cancer. Front. Oncol. 7, 85 (2017).
Lo, S. C., Li, X., Henzl, M. T., Beamer, L. J. & Hannink, M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 25, 3605–3617 (2006).
Koren, I. et al. The eukaryotic proteome is shaped by E3 ubiquitin ligases targeting C-terminal degrons. Cell 173, 1622–1635 (2018).
Lin, H. C. et al. C-terminal end-directed protein elimination by CRL2 ubiquitin ligases. Mol. Cell 70, 602–613 (2018). This reference and Koren et al. (2018) are the first studies to systematically investigate protein and peptide stability at scale,revealing that the extreme C termini of proteins can serve as necessary and sufficient degrons to trigger protein degradation.
Piatkov, K. I., Colnaghi, L., Békés, M., Varshavsky, A. & Huang, T. T. The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol. Cell 48, 926–933 (2012).
Ichikawa, S. et al. The E3 ligase adapter cereblon targets the C-terminal cyclic imide degron. Nature 610, 775–782 (2022).
Muhar, M. F. et al. C-terminal amides mark proteins for degradation via SCF–FBXO31. Nature 638, 519–527 (2025).
Zhang, Z. et al. Elucidation of E3 ubiquitin ligase specificity through proteome-wide internal degron mapping. Mol. Cell 83, 3377–3392 (2023).
Kussie, P. H. et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274, 948–953 (1996).
Zhou, M. et al. Molecular insights into degron recognition by CRL5. Nat. Commun. 15, 6177 (2024).
Liang, X. et al. A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures. Nat. Chem. Biol. 18, 1214–1223 (2022).
Luptak, J. et al. TRIM7 restricts coxsackievirus and norovirus infection by detecting the C-terminal glutamine generated by 3C protease processing. Viruses 14, 1610 (2022).
Ru, Y. et al. C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7. Proc. Natl Acad. Sci. USA 119, e2203218119 (2022).
Yang, S. W. et al. Structural basis for substrate recognition and chemical inhibition of oncogenic MAGE ubiquitin ligases. Nat. Commun. 11, 4931 (2020).
Grabarczyk, D. B. et al. Architecture of the UBR4 complex, a giant E4 ligase central to eukaryotic protein quality control. Science 389, 909–914 (2025).
Haakonsen, D. L. et al. Stress response silencing by an E3 ligase mutated in neurodegeneration. Nature 626, 874–880 (2024).
Santelli, E. et al. Structural analysis of Siah1–Siah-interacting protein interactions and insights into the assembly of an E3 ligase multiprotein complex. J. Biol. Chem. 280, 34278–34287 (2005).
Zeng, Z. et al. Structural basis of selective ubiquitination of TRF1 by SCFFbx4. Dev. Cell 18, 214–225 (2010).
Xing, W. et al. SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68 (2013).
Hu, Y. et al. Dynamic molecular architecture and substrate recruitment of cullin3–RING E3 ligase CRL3. Nat. Struct. Mol. Biol. 31, 336–350 (2024).
Jiang, W., Wang, W., Kong, Y. & Zheng, S. Structural basis for the ubiquitination of G protein βγ subunits by KCTD5/cullin3 E3 ligase. Sci. Adv. 9, eadg8369 (2023). This study reveals how a globular subunit of G proteins is recognized by a CRL3 substrate receptor to modulate and attenuate GPCR signaling.
Nguyen, D. M. et al. Structure and dynamics of a pentameric KCTD5/CUL3/Gβγ E3 ubiquitin ligase complex. Proc. Natl Acad. Sci. USA 121, e2315018121 (2024).
Xu, P. et al. The CRL5–SPSB3 ubiquitin ligase targets nuclear cGAS for degradation. Nature 627, 873–879 (2024).
Filippakopoulos, P. et al. Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4. J. Mol. Biol. 401, 389–402 (2010).
Cao, S. et al. Recognition of BACH1 quaternary structure degrons by two F-box proteins under oxidative stress. Cell 187, 7568–7584 (2024).
Goretzki, B. et al. Dual BACH1 regulation by complementary SCF-type E3 ligases. Cell 187, 7585–7602 (2024). This reference and Cao et al. (2024) provide mechanistic insights into how E3 ligases detect not only quaternary structure degrons but also the strength of interaction between protomers as a signal for protein degradation.
Lumpkin, R. J., Baker, R. W., Leschziner, A. E. & Komives, E. A. Structure and dynamics of the ASB9 CUL–RING E3 ligase. Nat. Commun. 11, 2866 (2020).
Teng, F. et al. Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme. Structure 31, 1431–1440 (2023).
Gottemukkala, K. V. et al. Non-canonical substrate recognition by the human WDR26–CTLH E3 ligase regulates prodrug metabolism. Mol. Cell 84, 1948–1963 (2024).
Sherpa, D. et al. GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme. Mol. Cell 81, 2445–2459 (2021).
Jenkyn-Bedford, M. et al. A conserved mechanism for regulating replisome disassembly in eukaryotes. Nature 600, 743–747 (2021). This report reveals how E3 ligases selectively ubiquitinate and disassemble the replisome complex only after the termination of DNA replication.
Grothusen, G. P. et al. DCAF15 control of cohesin dynamics sustains acute myeloid leukemia. Nat. Commun. 15, 5604 (2024).
Welcker, M. et al. Two diphosphorylated degrons control c-Myc degradation by the Fbw7 tumor suppressor. Sci. Adv. 8, eabl7872 (2022). This study demonstrates how two double-phosphorylated degrons in an important oncogene product are simultaneously engaged by a dimeric E3 ligase to initiate protein degradation.
Zhang, S., Tischer, T. & Barford, D. Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C. Nat. Commun. 10, 3863 (2019).
Kuchay, S. et al. FBXL2- and PTPL1-mediated degradation of p110-free p85β regulatory subunit controls the PI3K signalling cascade. Nat. Cell Biol. 15, 472–480 (2013).
Liu, Y. et al. Structural basis for the regulatory role of the PPxY motifs in the thioredoxin-interacting protein TXNIP. Biochem. J. 473, 179–187 (2016).
Rusnac, D. V. et al. Recognition of the diglycine C-end degron by CRL2KLHDC2 ubiquitin ligase. Mol. Cell 72, 813–822 (2018).
Düring, J. et al. Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1. Nat. Struct. Mol. Biol. 31, 364–377 (2024). This study provides the structural basis for how ligand-induced conformational changes are recognized by E3 ligases and how the autoinhibited state of a HECT E3 ligase is maintained.
Tsai, J. M. et al. UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability. Mol. Cell 83, 2753–2767 (2023).
Mark, K. G. et al. Orphan quality control shapes network dynamics and gene expression. Cell 186, 3460–3475 (2023).
Zeng, J. et al. Target-induced clustering activates Trim-Away of pathogens and proteins. Nat. Struct. Mol. Biol. 28, 278–289 (2021).
Benn, J. et al. Aggregate-selective removal of pathological tau by clustering-activated degraders. Science 385, 1009–1016 (2024).
Miller, L. V. C. et al. Co-opting templated aggregation to degrade pathogenic tau assemblies and improve motor function. Cell 187, 5967–5980 (2024).
Kokic, G., Wagner, F. R., Chernev, A., Urlaub, H. & Cramer, P. Structural basis of human transcription–DNA repair coupling. Nature 598, 368–372 (2021).
Matsuo, Y. et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 8, 159 (2017).
Grabarczyk, D. B. et al. HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain. Nat. Chem. Biol. 17, 1084–1092 (2021).
Hunkeler, M. et al. Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition. Mol. Cell 81, 3468–3480 (2021).
Meek, D. W. & Knippschild, U. Posttranslational modification of MDM2. Mol. Cancer Res. 1, 1017–1026 (2003).
Dou, H. et al. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 19, 184–192 (2012).
Gladkova, C., Maslen, S. L., Skehel, J. M. & Komander, D. Mechanism of parkin activation by PINK1. Nature 559, 410–414 (2018).
DaRosa, P. A. et al. Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature 517, 223–226 (2015).
Kuchay, S. et al. GGTase3 is a newly identified geranylgeranyltransferase targeting a ubiquitin ligase. Nat. Struct. Mol. Biol. 26, 628–636 (2019).
Bhat, S.A. et al. Geranylgeranylated SCFFBXO10 regulates selective outer mitochondrial membrane proteostasis and function. Cell Rep. 43, 114783 (2024).
Wang, H. et al. FBXL5 regulates IRP2 stability in iron homeostasis via an oxygen-responsive [2Fe2S] cluster. Mol. Cell 78, 31–41 (2020).
Ahel, J. et al. Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism. eLife 9, e56185 (2020). This study reveals a unique large E3 ligase containing an unusual dynein-like core with ATPase activity.
Crespillo-Casado, A. et al. Recognition of phylogenetically diverse pathogens through enzymatically amplified recruitment of RNF213. EMBO Rep. 25, 4979–5005 (2024).
Ito, S., Song, Y. H. & Imaizumi, T. LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol. Plant 5, 573–582 (2012).
Kwon, E. et al. Structural analysis of the regulation of blue-light receptors by GIGANTEA. Cell Rep. 39, 110700 (2022).
Alfieri, C. et al. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature 536, 431–436 (2016).
Manford, A. G. et al. Structural basis and regulation of the reductive stress response. Cell 184, 5375–5390 (2021). This study demonstrates that zinc ions act as molecular glues at the substrate–E3 interface, facilitating substrate degradation.
Scott, D. C. et al. E3 ligase autoinhibition by C-degron mimicry maintains C-degron substrate fidelity. Mol. Cell 83, 770–786 (2023).
Komatsu, M. et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12, 213–223 (2010).
Ehrmann, J. F. et al. Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex. Science 379, 1117–1123 (2023).
Hunkeler, M., Jin, C. Y. & Fischer, E. S. Structures of BIRC6–client complexes provide a mechanism of SMAC-mediated release of caspases. Science 379, 1105–1111 (2023).
Liu, S. S. et al. Molecular mechanisms underlying the BIRC6-mediated regulation of apoptosis and autophagy. Nat. Commun. 15, 891 (2024).
Peschard, P. et al. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol. Cell 27, 474–485 (2007).
Kozlov, G. et al. Structural basis for UBA-mediated dimerization of c-Cbl ubiquitin ligase. J. Biol. Chem. 282, 27547–27555 (2007).
Chen, X. et al. Mechanism of Ψ-Pro/C-degron recognition by the CRL2. Nat. Commun. 15, 3558 (2024).
Duda, D. M. et al. Structure of HHARI, a RING–IBR–RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure 21, 1030–1041 (2013).
Kostrhon, S. et al. CUL5–ARIH2 E3–E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation. Nat. Chem. Biol. 17, 1075–1083 (2021).
Scott, D. C. et al. Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell 166, 1198–1214 (2016).
Tan, X. et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 (2007).
Sheard, L. B. et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 468, 400–405 (2010).
McMinimy, R. et al. Reactive oxygen species control protein degradation at the mitochondrial import gate. Mol. Cell 84, 4612–4628 (2024).
Varshney, N. et al. A review of von Hippel–Lindau syndrome. J. Kidney Cancer VHL 4, 20–29 (2017).
Fischer, E. S. et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147, 1024–1039 (2011).
Johnson-Kerner, B. L., Roth, L., Greene, J. P., Wichterle, H. & Sproule, D. M. Giant axonal neuropathy: an updated perspective on its pathology and pathogenesis. Muscle Nerve 50, 467–476 (2014).
Ge, Z. et al. Integrated genomic analysis of the ubiquitin pathway across cancer types. Cell Rep. 23, 213–226 (2018).
Welcker, M. & Clurman, B. E. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 (2008).
Cuneo, M. J., O’Flynn, B. G., Lo, Y. H., Sabri, N. & Mittag, T. Higher-order SPOP assembly reveals a basis for cancer mutant dysregulation. Mol. Cell 83, 731–745 (2023).
Cerami, E. et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Schumacher, F. R., Sorrell, F. J., Alessi, D. R., Bullock, A. N. & Kurz, T. Structural and biochemical characterization of the KLHL3–WNK kinase interaction important in blood pressure regulation. Biochem. J. 460, 237–246 (2014).
Boyden, L. M. et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482, 98–102 (2012).
Tokheim, C. et al. Systematic characterization of mutations altering protein degradation in human cancers. Mol. Cell 81, 1292–1308 (2021).
Lin, Z. et al. Stabilizing mutations of KLHL24 ubiquitin ligase cause loss of keratin 14 and human skin fragility. Nat. Genet. 48, 1508–1516 (2016).
Northcott, P. A. et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017).
Chen, Z. et al. Disease-associated KBTBD4 mutations in medulloblastoma elicit neomorphic ubiquitylation activity to promote CoREST degradation. Cell Death Differ. 29, 1955–1969 (2022).
Xie, X. et al. Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations. Nature 639, 241–249 (2025).
Chen, Z. et al. Structural mimicry of UM171 and neomorphic cancer mutants co-opts E3 ligase KBTBD4 for HDAC1/2 recruitment. Nat. Commun. 16, 3144 (2025). This reference, Xie et al. (2025) and Yeo (2025) demonstrate how cancer associated mutations in an E3 ligase mimic the mechanism of action of a small molecule to induce the degradation of neosubstrates.
Mercuri, E., Sumner, C. J., Muntoni, F., Darras, B. T. & Finkel, R. S. Spinal muscular atrophy. Nat. Rev. Dis. Primers 8, 52 (2022).
Cho, S. & Dreyfuss, G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 24, 438–442 (2010).
Oleinikovas, V., Gainza, P., Ryckmans, T., Fasching, B. & Thomä, N. H. From thalidomide to rational molecular glue design for targeted protein degradation. Annu. Rev. Pharmacol. Toxicol. 64, 291–312 (2023).
Yoon, H., Rutter, J. C., Li, Y. D. & Ebert, B. L. Induced protein degradation for therapeutics: past, present, and future. J. Clin. Invest. 134, e175265 (2024).
Konstantinidou, M. & Arkin, M. R. Molecular glues for protein–protein interactions: progressing toward a new dream. Cell Chem. Biol. 31, 1064–1088 (2024).
Wang, B., Cao, S. & Zheng, N. Emerging strategies for prospective discovery of molecular glue degraders. Curr. Opin. Struct. Biol. 86, 102811 (2024).
Békés, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200 (2022).
Lu, P. et al. Selective degradation of multimeric proteins by TRIM21-based molecular glue and PROTAC degraders. Cell 187, 7126–7142 (2024). The authors of this study discovered a novel MGD that targets the nuclear pore complex and designed PROTAC molecules that selectively target multimeric proteins.
Zhuang, Z. et al. Charged molecular glue discovery enabled by targeted degron display. Preprint at bioRxiv https://doi.org/10.1101/2024.09.24.614843 (2024).
Yeo, M. J. R. UM171 glues asymmetric CRL3–HDAC1/2 assembly to degrade CoREST corepressors. Nature 639, 232–240 (2025).
Hanzl, A. et al. Primed for degradation: how weak protein interactions enable molecular glue degraders. Curr. Opin. Struct. Biol. 92, 103052 (2025).
Simonetta, K. R. et al. Prospective discovery of small molecule enhancers of an E3 ligase–substrate interaction. Nat. Commun. 10, 1402 (2019).
Petzold, G., Fischer, E. S. & Thomä, N. H. Structural basis of lenalidomide-induced CK1α degradation by the CRL4CRBN ubiquitin ligase. Nature 532, 127–130 (2016).
Matyskiela, M. E. et al. A novel cereblon modulator recruits GSPT1 to the CRL4CRBN ubiquitin ligase. Nature 535, 252–257 (2016).
Wing, C. E. et al. Allosteric degraders induce CRL5ASB8 mediated degradation of XPO1. Preprint at bioRxiv https://doi.org/10.1101/2024.10.07.617049 (2024).
Song, K. W. et al. RTK-dependent inducible degradation of mutant PI3Kα drives GDC-0077 (inavolisib) efficacy. Cancer Discov. 12, 204–219 (2022).
Ferretti, S. et al. Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers. Nature 629, 443–449 (2024).
Hennes, E. et al. Monovalent pseudo-natural product degraders supercharge the native degradation of IDO1 by KLHDC3. Preprint at bioRxiv https://doi.org/10.1101/2024.07.10.602857 (2024).
Boike, L. et al. Discovery of a functional covalent ligand targeting an intrinsically disordered cysteine within MYC. Cell Chem. Biol. 28, 4–13 (2021).
Gowans, F. A. Covalent degrader of the oncogenic transcription factor β-catenin. J. Am. Chem. Soc. 146, 16856–16865 (2024).
Roy, N. et al. Suppression of NRF2-dependent cancer growth by a covalent allosteric molecular glue. Preprint at bioRxiv https://doi.org/10.1101/2024.10.04.616592 (2024).
Sauvé, V. et al. Activation of parkin by a molecular glue. Nat. Commun. 15, 7707 (2024).
Wang, W. et al. MDM2 inhibitors for cancer therapy: the past, present, and future. Pharm. Rev. 76, 414–453 (2024).
Crisman, E. et al. KEAP1–NRF2 protein–protein interaction inhibitors: design, pharmacological properties and therapeutic potential. Med. Res. Rev. 43, 237–287 (2023).
Kimani, S. W. et al. The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition. Commun. Biol. 6, 1272 (2023).
Collins, G. P. et al. A first-in-human phase 1 trial of NX-1607, a first-in-class oral CBL-B inhibitor, in patients with advanced malignancies including DLBCL. Blood 142, 3093 (2023).
Rothman, A. M. K. et al. Therapeutic potential of allosteric HECT E3 ligase inhibition. Cell 188, 2603–2620 (2025).
Timms, R. T. et al. A glycine-specific N-degron pathway mediates the quality control of protein. Science 365, eaaw4912 (2019).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Zhou, G. et al. An artificial intelligence accelerated virtual screening platform for drug discovery. Nat. Commun. 15, 7761 (2024).
Cao, S. et al. Defining molecular glues with a dual-nanobody cannabidiol sensor. Nat. Commun. 13, 815 (2022).
Petzold, G. et al. Mining the CRBN target space redefines rules for molecular glue-induced neosubstrate recognition. Science 389, eadt6736 (2025).
Harper, J. W. & Schulman, B. A. Cullin–RING ubiquitin ligase regulatory circuits: a quarter century beyond the F-box hypothesis. Annu. Rev. Biochem. 90, 403–429 (2021).
Hopf, L. V. M. et al. Structure of CRL7FBXW8 reveals coupling with CUL1–RBX1/ROC1 for multi-cullin–RING E3-catalyzed ubiquitin ligation. Nat. Struct. Mol. Biol. 29, 854–862 (2022).
Horn-Ghetko, D. et al. Noncanonical assembly, neddylation and chimeric cullin–RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex. Nat. Struct. Mol. Biol. 31, 1083–1094 (2024).
Barford, D. Structural interconversions of the anaphase-promoting complex/cyclosome (APC/C) regulate cell cycle transitions. Curr. Opin. Struct. Biol. 61, 86–97 (2020).
Qiao, S. et al. Interconversion between anticipatory and active GID E3 ubiquitin ligase conformations via metabolically driven substrate receptor assembly. Mol. Cell 77, 150–163 (2020).
Sakamoto, K. M. et al. Protacs: chimeric molecules that target proteins to the Skp1–cullin–F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. USA 98, 8554–8559 (2001).
Acknowledgements
B.W. and N.Z. are supported by the Howard Hughes Medical Institute, the Washington Research Foundation and funds from the US National Institutes of Health (R01 HD097408, R01 DA056370 and R01HL112808). N.Z. is a Howard Hughes Medical Institute Investigator.
Author information
Authors and Affiliations
Contributions
B.W. and N.Z. conceived the central themes of the Review. B.W. wrote the initial draft. N.Z. made the figures. B.W. and N.Z. completed the final preparation.
Corresponding authors
Ethics declarations
Competing interests
N.Z. is one of the scientific cofounders and a shareholder of SEED Therapeutics, and serves as a member of the scientific advisory boards of SyntheX, Molecular Glue Labs, Cold Start Therapeutics and Differentiated Therapeutics, with financial interests. B.W. declares no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Chao Xu, Cheng Dong and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
References for Supplementary Table 1
Supplementary Table 1
Structurally characterized human E3–degron complexes.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, B., Zheng, N. Degrons and degradation signals beyond short linear motifs. Nat Chem Biol (2025). https://doi.org/10.1038/s41589-025-02056-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41589-025-02056-2