Abstract
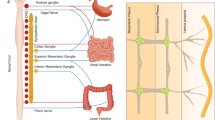
The nervous and immune systems cooperate to regulate mucosal barrier integrity. Nevertheless, whether enteric neurons establish neuroepithelial interactions to coordinate immunity remains elusive. Here, we identified neuroepithelial interactions that differentially control intestinal type 1 and type 2 immunity. Gut epithelial cells expressed vasoactive intestinal peptide (VIP) receptor 1 (VIPR1), and chemogenetic modulation of enteric VIPergic neurons led to altered epithelial-derived cytokines. Epithelial-intrinsic deletion of Vipr1 resulted in diminished type 1 immunity, including reduced type 1 alarmins and intraepithelial lymphocytes. In contrast, epithelial Vipr1 deficiency led to enhanced type 2 immunity, comprising increased type 2 alarmins, tuft cells and activated group 2 innate lymphoid cells. Disruption of neuroepithelial VIP–VIPR1 interactions resulted in increased susceptibility to invasive bacterial infection, which contrasted with enhanced resistance to parasite infection. Our work identifies a multi-tissue axis that controls type 1 and type 2 immunity, deciphering how neuroepithelial interactions distinctively set gut immunity programs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The datasets generated in this study are available from the corresponding author upon request. Bulk RNA-seq and single-cell RNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession numbers GSE308754 and GSE308755, respectively. Source data are provided with this paper.
References
Peterson, L. W. & Artis, D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153 (2014).
Lockhart, A., Mucida, D. & Bilate, A. M. Intraepithelial lymphocytes of the intestine. Annu Rev. Immunol. 42, 289–316 (2024).
Lodolce, J. P. et al. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 9, 669–676 (1998).
Spits, H. & Mjosberg, J. Heterogeneity of type 2 innate lymphoid cells. Nat. Rev. Immunol. 22, 701–712 (2022).
von Moltke, J., Ji, M., Liang, H. E. & Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016).
Godinho-Silva, C., Rendas, M. & Veiga-Fernandes, H. Neuro-ILC interactions in host physiology and defence. Eur. J. Immunol. 55, e70037 (2025).
Klein Wolterink, R. G. J., Wu, G. S., Chiu, I. M. & Veiga-Fernandes, H. Neuroimmune interactions in peripheral organs. Annu. Rev. Neurosci. 45, 339–360 (2022).
Klose, C. S. N. & Veiga-Fernandes, H. Neuroimmune interactions in peripheral tissues. Eur. J. Immunol. 51, 1602–1614 (2021).
Pirzgalska, R. M. & Veiga-Fernandes, H. Type 2 neuroimmune circuits in the shaping of physiology. Immunity 56, 695–703 (2023).
Sestan, M. et al. Neuronal–ILC2 interactions regulate pancreatic glucagon and glucose homeostasis. Science 387, eadi3624 (2025).
Moriyama, S. et al. β 2-adrenergic receptor-mediated negative regulation of group 2 innate lymphoid cell responses. Science 359, 1056–1061 (2018).
Uddin, J. et al. CGRP-related neuropeptide adrenomedullin 2 promotes tissue-protective ILC2 responses and limits intestinal inflammation. Nat. Immunol. 26, 1516–1526 (2025).
Talbot, S. et al. Silencing nociceptor neurons reduces allergic airway inflammation. Neuron 87, 341–354 (2015).
Nussbaum, J. C. et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248 (2013).
Pascal, M. et al. The neuropeptide VIP potentiates intestinal innate type 2 and type 3 immunity in response to feeding. Mucosal Immunol. 15, 629–641 (2022).
Talbot, J. et al. Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature 579, 575–580 (2020).
Seillet, C. et al. The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat. Immunol. 21, 168–177 (2020).
Lei, C. et al. Enteric VIP-producing neurons maintain gut microbiota homeostasis through regulating epithelium fucosylation. Cell Host Microbe 30, 1417–1434.e1418 (2022).
Schwerdtfeger, L. A. & Tobet, S. A. Vasoactive intestinal peptide regulates ileal goblet cell production in mice. Physiol. Rep. 8, e14363 (2020).
Wu, X. et al. Vasoactive intestinal polypeptide promotes intestinal barrier homeostasis and protection against colitis in mice. PLoS ONE 10, e0125225 (2015).
Ericsson, A. C. et al. The G protein-coupled receptor, VPAC1, mediates vasoactive intestinal peptide-dependent functional homeostasis of the gut microbiota. Gastro. Hep. Adv. 1, 253–264 (2022).
Morarach, K. et al. Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing. Nat. Neurosci. 24, 34–46 (2021).
Drokhlyansky, E. et al. The human and mouse enteric nervous system at single-cell resolution. Cell 182, 1606–1622.e1623 (2020).
Moody, T. W., Ito, T., Osefo, N. & Jensen, R. T. VIP and PACAP: recent insights into their functions/roles in physiology and disease from molecular and genetic studies. Curr. Opin. Endocrinol. Diabetes Obes. 18, 61–67 (2011).
Timmermans, S., Souffriau, J. & Libert, C. A general introduction to glucocorticoid biology. Front. Immunol. 10, 1545 (2019).
Godinho-Silva, C. et al. Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature 574, 254–258 (2019).
Shimba, A. et al. Glucocorticoids drive diurnal oscillations in T cell distribution and responses by inducing interleukin-7 receptor and CXCR4. Immunity 48, 286–298 e286 (2018).
Lin, X. et al. IL-17RA-signaling in Lgr5(+) intestinal stem cells induces expression of transcription factor ATOH1 to promote secretory cell lineage commitment. Immunity 55, 237–253.e238 (2022).
Schneider, C. et al. A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell 174, 271–284.e214 (2018).
Nadjsombati, M. S. et al. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49, 33–41.e37 (2018).
Veiga-Fernandes, H. & Mucida, D. Neuro-immune interactions at barrier surfaces. Cell 165, 801–811 (2016).
Larsson, L. I. et al. Localization of vasoactive intestinal polypeptide (VIP) to central and peripheral neurons. PNAS 73, 3197–3200 (1976).
Costa, M. & Furness, J. B. The origins, pathways and terminations of neurons with VIP-like immunoreactivity in the guinea-pig small intestine. Neuroscience 8, 665–676 (1983).
Iwasaki, M., Akiba, Y. & Kaunitz, J. D. Recent advances in vasoactive intestinal peptide physiology and pathophysiology: focus on the gastrointestinal system. F1000Res https://doi.org/10.12688/f1000research.18039.1 (2019).
Tuganbaev, T. et al. Diet diurnally regulates small intestinal microbiome-epithelial-immune homeostasis and enteritis. Cell 182, 1441–1459.e1421 (2020).
Loh, D. H., Abad, C., Colwell, C. S. & Waschek, J. A. Vasoactive intestinal peptide is critical for circadian regulation of glucocorticoids. Neuroendocrinology 88, 246–255 (2008).
Fabricius, D. et al. Characterization of intestinal and pancreatic dysfunction in VPAC1-null mutant mouse. Pancreas 40, 861–871 (2011).
Lelievre, V. et al. Gastrointestinal dysfunction in mice with a targeted mutation in the gene encoding vasoactive intestinal polypeptide: a model for the study of intestinal ileus and Hirschsprung’s disease. Peptides 28, 1688–1699 (2007).
Gerbe, F. et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J. Cell Biol. 192, 767–780 (2011).
Lennicke, C. et al. Loss of epithelium-specific GPx2 results in aberrant cell fate decisions during intestinal differentiation. Oncotarget 9, 539–552 (2018).
Jin, S. et al. Inference and analysis of cell–cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Stoklasek, T. A., Schluns, K. S. & Lefrancois, L. Combined IL-15/IL-15Rα immunotherapy maximizes IL-15 activity in vivo. J. Immunol. 177, 6072–6080 (2006).
Ma, L. J., Acero, L. F., Zal, T. & Schluns, K. S. Trans-presentation of IL-15 by intestinal epithelial cells drives development of CD8αα IELs. J. Immunol. 183, 1044–1054 (2009).
Chawla, A. S. et al. Distinct cell death pathways induced by granzymes collectively protect against intestinal Salmonella infection. Mucosal Immunol. 17, 1242–1255 (2024).
Hoytema van Konijnenburg, D. P. et al. Intestinal epithelial and intraepithelial T cell crosstalk mediates a dynamic response to infection. Cell 171, 783–794.e713 (2017).
Helou, D. G. et al. PD-1 pathway regulates ILC2 metabolism and PD-1 agonist treatment ameliorates airway hyperreactivity. Nat. Commun. 11, 3998 (2020).
Wang, Y. et al. Bi-directional communication between intrinsic enteric neurons and ILC2s inhibits host defense against helminth infection. Immunity 58, 465–480.e468 (2025).
Barilla, R. M. et al. Type 2 cytokines act on enteric sensory neurons to regulate neuropeptide-driven host defense. Science 389, 260–267 (2025).
Feng, X. et al. Tuft cell IL-17RB restrains IL-25 bioavailability and reveals context-dependent ILC2 hypoproliferation. Nat. Immunol. 26, 567–581 (2025).
He, S. et al. Gut intraepithelial T cells calibrate metabolism and accelerate cardiovascular disease. Nature 566, 115–119 (2019).
Sullivan, Z. A. et al. γδT cells regulate the intestinal response to nutrient sensing. Science https://doi.org/10.1126/science.aba8310 (2021).
Veiga-Fernandes, H. & Freitas, A. A. The S(c)ensory immune system theory. Trends Immunol. https://doi.org/10.1016/j.it.2017.02.007 (2017).
Madison, B. B. et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 277, 33275–33283 (2002).
el Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Taniguchi, H. et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013 (2011).
Mittelstadt, P. R., Monteiro, J. P. & Ashwell, J. D. Thymocyte responsiveness to endogenous glucocorticoids is required for immunological fitness. J. Clin. Investig. 122, 2384–2394 (2012).
Sciolino, N. R. et al. Recombinase-dependent mouse lines for chemogenetic activation of genetically defined cell types. Cell Rep. 15, 2563–2573 (2016).
Zhu, H. et al. Cre-dependent DREADD (designer receptors exclusively activated by designer drugs) mice. Genesis 54, 439–446 (2016).
Mombaerts, P. et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992).
Maekawa, Y. et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat. Immunol. 9, 1140–1147 (2008).
Cardoso, V. et al. Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature 549, 277–281 (2017).
Ibiza, S. et al. Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature 535, 440–443 (2016).
Cardoso, F. et al. Neuro-mesenchymal units control ILC2 and obesity via a brain-adipose circuit. Nature 597, 410–414 (2021).
Fonseca-Pereira, D. et al. The neurotrophic factor receptor RET drives haematopoietic stem cell survival and function. Nature 514, 98–101 (2014).
van de Pavert, S. A. et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature 508, 123–127 (2014).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Choudhary, S. & Satija, R. Comparison and evaluation of statistical error models for scRNA-seq. Genome Biol. 23, 27 (2022).
Jin, S., Plikus, M. V. & Nie, Q. CellChat for systematic analysis of cell–cell communication from single-cell transcriptomics. Nat. Protoc. 20, 180–219 (2025).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Harrison, P. W. et al. Ensembl 2024. Nucleic Acids Res. 52, D891–D899 (2024).
Matsumoto, K. et al. Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat. Protoc. 14, 3506–3537 (2019).
Obata, Y. & Pachnis, V. The effect of microbiota and the immune system on the development and organization of the enteric nervous system. Gastroenterology 151, 836–844 (2016).
Kugler, S., Kilic, E. & Bahr, M. Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 10, 337–347 (2003).
Rastelli, D. et al. Diminished androgen levels are linked to irritable bowel syndrome and cause bowel dysfunction in mice. J. Clin. Invest. https://doi.org/10.1172/JCI150789 (2022).
Muller, P. A. et al. Microbiota modulate sympathetic neurons via a gut–brain circuit. Nature 583, 441–446 (2020).
Klose, C. S. et al. The transcription factor T-bet is induced by IL-15 and thymic agonist selection and controls CD8αα(+) intraepithelial lymphocyte development. Immunity 41, 230–243 (2014).
Balbontin, R., Vlamakis, H. & Kolter, R. Mutualistic interaction between Salmonella enterica and Aspergillus niger and its effects on Zea mays colonization. Microb. Biotechnol. 7, 589–600 (2014).
Bouchery, T. et al. ILC2s and T cells cooperate to ensure maintenance of M2 macrophages for lung immunity against hookworms. Nat. Commun. 6, 6970 (2015).
Erben, U. et al. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol. 7, 4557–4576 (2014).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. Cell detection with star-convex polygons. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, 265–273 (Springer International Publishing, 2018).
Acknowledgements
We thank the Vivarium, Flow Cytometry, Histopathology, Advanced BioImaging and BioOptics Experimental Platform, Molecular Biology, Hardware and Software Platform, Glass Wash, and Media platforms at Champalimaud Foundation. We thank Congento LISBOA-01-0145-FEDER-022170. We thank Mariana Monteiro (Histopathology platform, Champalimaud Foundation) for quantification of goblet, tuft and Ki67+ cells. We thank M. Patrício and S. Tehrani (Champalimaud Foundation, Portugal) for technical help with N. brasiliensis larvae. S. entericaserovar Typhimurium 14028 was kindly provided by I. Gordo, GIMM, Portugal. Infective (iL3) worms of N. brasiliensis were kindly provided by J. Allen, University of Manchester, UK. We thank C. Schneider (University of Zurich, Switzerland), M. Rao (Harvard Medical School, USA), P. Bastos, M. Martinez-Lopez, A. Rasteiro and M. Aliseychik (Champalimaud Foundation, Portugal) for helpful discussions. R.M.P. was supported by FCT (CEECIND/03601/2018; PTDC/MED-IMU/6381/2020), European Crohn’s and Colitis Organisation Grant, the European Foundation for the Study of Diabetes (EFSD)/Lilly Young Investigator award, EFSD/Novo Nordisk (NN) Rising Star Fellowship and the European Association for the Study of Obesity/NN grant. J.R. was supported by EU HORIZON-MISS-2021-CANCER-02-03 (GENIAL 101096312). C.G.-S. was supported FCT (2023.07506.CEECIND and PTDC/MED-IMU/2189/2021). M.R. was supported by FCT (PTDC/MED-IMU/2189/2021). C.W. was supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) under Germany’s Excellence Strategy – EXC2151 – 390873048 and WI 4554/6-1. C.S.N.K. was funded by DFG, under Germany’s Excellence Strategy – EXC 3118/1 – project 533770413. V.M.L was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases. M.F. and J.T. were supported by la Caixa Foundation (LCF/PR/HR22/52420007) and FCT (2022.06145.PTDC). C.M.M. and V.C. were supported by the European Research Council (101116335) and by the European Haematology Association. H.V.-F. was supported by the European Research Council (647274 and 101097830); Paul G. Allen Frontiers Group (12826); Chan Zuckerberg Initiative (INFL-0000000193); La Caixa (HR20-00841); EU HORIZON-MISS-2021-CANCER-02-03 (GENIAL 101096312) and FCT (PTDC/MED-IMU/6653/2020).
Author information
Authors and Affiliations
Contributions
Initial observations leading to the study: R.M.P.; conceptualization: R.M.P. and H.V.-F.; methodology: R.M.P., B.H.-A., B.R., E.d.S., J.T., C.G.-S., M.R., T.C., V.C., R.R.-T., C.W., C.S.N.K., T.C., V.M.L., C.M.M., M.F. and H.V.-F.; investigation: R.M.P., B.H.-A., B.R., E.d.S., J.T., J.R., C.G.-S., M.R., M.P., I.G., V.C., M.O.J. and P.M.F.; visualization: R.M.P., B.H.-A., E.d.S. and T.C.; funding acquisition: R.M.P. and H.V.-F.; project administration: H.R. and H.V.-F.; supervision: R.M.P. and H.V.-F.; and writing: R.M.P. and H.V.-F.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: L. A. Dempsey, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
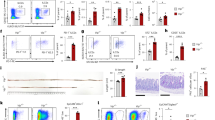
Extended Data Fig. 1 Epithelia-autonomous glucocorticoid signals are dispensable to IELs and ILC2 in the small intestine.
(a–c) Flow cytometry gating strategy. a, small intestinal epithelial cells. b, small intestinal IELs. c, small intestinal ILC2s in the lamina propria (d, e) Flow cytometry validation of ILC gating strategy with transcription factors. d, GATA2 for ILC2. e, RORγt for ILC3. f, Flow cytometry analysis of small intestinal IELs from Nr3c1fl (n = 7) and Nr3c1∆Villin (n = 6) mice. g, Flow cytometry analysis of small intestinal ILC2s from the lamina propria of Nr3c1fl (n = 5) and Nr3c1∆Villin (n = 5) mice. (f, g) Mean and error bars: s.e.m. n represents biologically independent animals. (f, g) Two-tailed unpaired t test with Welch correction.
Extended Data Fig. 2 VIPergic modulation of epithelial-derived alarmins.
a, Representative images of CD3+cells in the duodenum obtained by confocal microscopy. Scale bar, 20μm. VIPergic neurons (red); CD3+cells (green). b, Representative plots from flow cytometry analysis of small intestinal lamina propria of R26-TomatoVIP mice. c, Relative expression of type 1 and type 2 alarmins in purified intestinal epithelial cells from the duodenum of R26-hM3Dqfl (n = 3) and R26-hM3DqVIP (n = 3) mice under steady-state conditions, without CNO administration. (d–f) Systemic chemogenetic manipulation of VIP-expressing cells. d, Relative expression of type 2 alarmins in purified epithelial cells from the duodenum of R26-hM3Dqfl (n = 8) and R26-hM3DqVIP (n = 8) mice following systemic CNO-driven chemogenetic activation. e, Glucocorticoid levels in the serum of R26-hM3Dqfl (n = 5) and R26-hM3DqVIP (n = 6) mice following CNO-driven chemogenetic activation. Right panel depicts representative images of spleens from R26-hM3Dqfl and R26-hM3DqVIP mice following CNO-driven chemogenetic activation. f, Relative expression of type 2 alarmins in purified epithelial cells from the duodenum of R26-hM4Difl (n = 8) and R26-hM4DiVIP (n = 8) mice following CNO-driven chemogenetic inhibition. g, Validation of infection efficiency in duodenal chemogenetic activation (top) and inhibition (bottom) by confocal microscopy of duodenal sections. Scale bar, 20μm. AAV9-targetting construct (magenta), VIPergic neurons (yellow), nuclei (DAPI, blue). h, VIP levels in the duodenum from Vip-Cre mice (Vip-Cre.AAV9-hM3DqGut n = 6, represented in magenta) and control littermates (WT.AAV9-hM3DqGut n = 6, represented in dark gray) injected with AAV9-carrying activatory DREADDSs and after CNO-driven activation. i, Relative expression of mouse Vipr1 and Vipr2 (Left) (n = 9) and human VIPR1 and VIPR2 (Right) (n = 12) in small intestinal epithelial cells. (c–f, h, i) Mean and error bars: s.e.m. n represents biologically independent animals or samples. (c–f, h, i) Two-tailed unpaired t test with Welch correction.
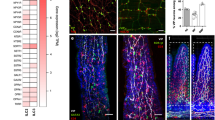
Extended Data Fig. 3 Vipr1 deletion in intestinal epithelial cells.
a, Graphical representation of Vipr1 gene targeting. Blue boxes represent exons; neo: neomycin resistance cassette; FRT: recognition sequence for flp recombinase; loxP: recognition sequence for Cre recombinase. b, Example of genotype PCR of Vipr1fl/fl mice. Primer sequences are provided in the Methods section. ntc-no template control. c, scRNA-seq analysis of Vipr1 and Vipr2 expression in small intestinal epithelial cells from Vipr1fl and Vipr1∆Villin mice. Data shown are representative of intestinal segments collected from two animals per group. d, Representative hematoxylin and eosin (H&E) staining of small intestinal tissue (duodenum-left; ileum-right) from Vipr1fl and Vipr1∆Villin mice. Original magnification 5x (upper row: scale bar, 200μm) and 40x (lower row: scale bar, 20μm). e, Ki-67 index in the duodenum (upper panel) and ileum (lower panel) from Vipr1fl (n = 6–7) and Vipr1∆Villin (n = 6) mice. f, Representative anti-Ki-67 staining in the duodenum (upper panels) and ileum (lower panels) from Vipr1fl and Vipr1∆Villin mice. Scale bar, 40μm (e) Mean and error bars: s.e.m. n represents biologically independent samples. Two-tailed unpaired t test with Welch correction.
Extended Data Fig. 4 Profiling in Vipr1 conditional knockout mice and tuft cell gating.
a, VIP levels in the small intestine (duodenum) from Vipr1fl (n = 6) and Vipr1∆Villin (n = 5) mice. b, VIP levels in serum from Vipr1fl (n = 5) and Vipr1∆Villin (n = 5) mice. c, Intestinal transit time measured in Vipr1fl (n = 6) and Vipr1∆Villin (n = 6) mice. d, scRNA-seq analysis of Cftr and Slc12a2 expression in small intestinal epithelial cells from Vipr1fl and Vipr1∆Villin mice. Data shown are representative of intestinal segments collected from two animals per group. e, Fecal water content measured as wet-to-dry weight in Vipr1fl (n = 6) and Vipr1∆Villin (n = 6) mice. f, Representative plots of flow cytometry gating for small intestinal tuft cells. g, Relative expression of Vipr1 in purified epithelial cells from the duodenum and ileum of Vipr1fl (n = 6) and Vipr1∆VillinERT2 (n = 5) mice following tamoxifen-driven recombination. Expression was assessed using primers targeting exon 2 of the Vipr1 gene. (a–c, e, g) Mean and error bars: s.e.m. n represents biologically independent animals. (a–c, e) Two-tailed unpaired t test with Welch correction. (d) Two-tailed Wilcoxon rank-sum test. n.d. not detected.
Extended Data Fig. 5 Epithelial-intrinsic Vipr1 deletion shape intestinal IELs and cell-intrinsic Vipr2 signaling is dispensable to IEL homeostasis.
a, Flow cytometry analysis of small intestinal IELs collected from the duodenum (duo) and ileum of Vipr1fl (n = 6) and Vipr1∆Villin (n = 4) mice after microbiota depletion with antibiotics. b, Flow cytometry analysis of small intestinal IELs collected from the duodenum (duo) and ileum of Vipr1fl (n = 5) and Vipr1∆VillinERT2 (n = 5) after antibiotic treatment. c, d, Flow cytometry analysis of small intestinal IELs collected from the duodenum (duo) and ileum of Vipr2fl (n = 5) and Vipr2∆CD8a (n = 5) mice. e, Relative expression of type 1 (Il15, Il18) and type 2 (Il33, Il25) alarmins in murine organoids stimulated with VIP or vehicle. Vehicle n = 5; VIP n = 5. (a–e) Mean and error bars: s.e.m. n represents biologically independent animals. (a–d) Ordinary one-way ANOVA with Šídák multiple comparisons test. (e) Two-tailed unpaired t test with Welch correction.
Extended Data Fig. 6 Neuroepithelial interactions modulate gut defense against bacteria.
(a–d) Co-housed Vipr1fl and Vipr1∆Villin mice were orally infected with Salmonella enterica. a, Relative expression of type 1 (Il15, Il18) and type 2 (Il33) alarmins in purified epithelial crypts from the duodenum of Vipr1fl (n = 5) and Vipr1∆Villin (n = 5) mice at day 1 post-infection. b, GFP immunostaining in Salmonella infected mice. Peyer’s Patches (PPs) from the Vipr1fl mice (left) displayed preserved morphological features of the follicle-associated epithelium (arrow) and lack of GFP+ staining, whereas PPs from Vipr1∆Villin mice showed numerous GFP+ Salmonella (brown), erosion and necrosis of the epithelium (asterisk; right panel). DAB counterstained with Harris’ hematoxylin. Original magnification: 40x, Scale bar, 40 µm (first two panels); 100x, Scale bar, 20 µm (magnified panels). c, Histopathology of the small intestine (ileum) at day 1 post-infection. Pathological changes include moderate to severe enteritis characterized by a mononuclear-rich inflammatory infiltrate in the mucosa (black arrows). Necrotic cell debris in the lumen (*) and necrosis of epithelial cells (open arrow); pronounced villous atrophy (open arrowhead) and villous blunting are more prominent in the Vipr1∆Villin mice. Hematoxylin and eosin (H&E). Scale bar, 200 μm (top); 25 μm (bottom). d, Histopathology of the small intestine (jejunum) at day 2 post-infection. Pathological changes in Vipr1∆Villin mice include mild enteritis (black arrows), abundant necrotic cell debris in the lumen (*), and necrosis of epithelial cells (white arrows). Hematoxylin and eosin (H&E). Scale bar, 400 μm (top); 50 μm (bottom). (a) Mean and error bars: s.e.m. n represents biologically independent animals. Two-tailed unpaired t test with Welch correction.
Extended Data Fig. 7 Epithelial-intrinsic Vipr1 deletion and intestinal immune cells.
a, Flow cytometry analysis of immune cell populations in the small intestinal lamina propria from Vipr1fl and Vipr1∆Villin mice. Absolute numbers of CD4 T cells, TCRαβ CD8 T cells, TCRγδ CD8 T cells, B cells, cDC1, cDC2, macrophages, and neutrophils were determined in Vipr1fl (n = 7) and Vipr1∆Villin (n = 6) mice, while ILC2 and ILC3 were analyzed in Vipr1fl (n = 11) and Vipr1∆Villin (n = 10) mice, and ILC1 and NK cells were assessed in Vipr1fl (n = 4) and Vipr1∆Villin (n = 4) mice. b, Flow cytometry gating strategy for small intestinal lamina propria immune cells. (a) Mean and error bars: s.e.m. n represents biologically independent animals. Two-tailed unpaired t test with Welch correction.
Extended Data Fig. 8 Epithelial-intrinsic Vipr1 deletion amplifies type 2 responses independently of microbiota.
(a–d) Flow cytometry analysis after microbiota depletion. a, Small intestinal PD-1+ ILC2 from the lamina propria of Vipr1fl (n = 6) and Vipr1∆Villin (n = 4) mice. b, Small intestinal eosinophils from the lamina propria of Vipr1fl (n = 6) and Vipr1∆Villin (n = 4). c, Small intestinal PD-1+ ILC2 from the lamina propria of Vipr1fl (n = 5) and Vipr1∆VillinERT2 (n = 5) mice. d, Small intestinal eosinophils from the lamina propria of Vipr1fl (n = 5) and Vipr1∆VillinERT2 (n = 6) mice. (e, f) Vipr2 deletion in IL-5–producing cells. e, Flow cytometry analysis of small intestinal ILC2 from the lamina propria of Vipr2WTIl5 (n = 4) and Vipr2∆Il5 (n = 5) mice. f, Flow cytometry analysis of cytokine production by small intestinal ILC2 from the lamina propria of Vipr2WTIl5 (n = 4) and Vipr2∆Il5 (n = 5) mice. g, Flow cytometry analysis of PD-1+ ILC2 from the mesenteric lymph nodes (mLN) from Vipr1fl (n = 6) and Vipr1∆Villin (n = 6). h, Small intestinal PD-1+ ILC2 and eosinophils isolated from the lamina propria after duodenal chemogenetic inhibition of VIPergic neurons. WT.AAV9-hM4DiGut n = 6; Vip-Cre.AAV9-hM4DiGut n = 5. i, Representative images of PAS staining in the ileum of Rag1−/−.Vipr1fl and Rag1−/−.Vipr1∆VillinERT2 mice. Periodic acid-Schiff (PAS). Scale bar, 200 μm (top panels) and 20 μm (bottom panels). j, Small intestinal PD-1+ ILC2 isolated from the lamina propria of Rag1−/−.Vipr1fl (n = 5) and Rag1−/−.Vipr1∆VillinERT2 (n = 4) mice after antibiotic treatment. k, Small intestinal eosinophils isolated from the lamina propria of Rag1−/−.Vipr1fl (n = 5) and Rag1−/−.Vipr1∆VillinERT2 (n = 4) mice after antibiotic treatment. (a–h, j, k) Mean and error bars: s.e.m. n represents biologically independent animals. (a, b) Two-tailed unpaired Mann–Whitney U-test. (c–g, j, k) Two-tailed unpaired t test with Welch correction. (h) Ordinary one-way ANOVA with Šídák multiple comparisons test.
Extended Data Fig. 9 Neuroepithelial interactions modulate gut defense against worm infections.
(a, b) Co-housed Vipr1fl and Vipr1∆Villin mice were infected with Nippostrongylus brasiliensis. a, Relative expression of type 1 (Il15, Il18) and type 2 (Il33) alarmins in purified epithelial crypts from the duodenum of Vipr1fl (n = 6) and Vipr1∆Villin (n = 5) mice at day 5 post-infection. b, Representative H&E images from the duodenum and ileum of the Vipr1fl and Vipr1∆Villin mice showing a mild increase in inflammatory cell infiltration in the mucosa of some villi (asterisks), predominantly composed of mononuclear cells. Hematoxylin and eosin (H&E). Scale bar, 200 μm (top panels) and 50 μm (bottom panels). (a) Mean and error bars: s.e.m. n represents biologically independent animals. Two-tailed unpaired Mann-Whitney U test.
Supplementary information
Supplementary Information
Unprocessed PCR gel from Extended Data Fig. 3b.
Source data
Source Data Fig. 1
Source Data Fig. 1.
Source Data Fig. 2
Source Data Fig. 2.
Source Data Fig. 3
Source Data Fig. 3.
Source Data Fig. 4
Source Data Fig. 4.
Source Data Fig. 5
Source Data Fig. 5.
Source Data Fig. 6
Source Data Fig. 6.
Source Data Fig. 7
Source Data Fig. 7.
Source Data Extended Data Fig. 1
Source Data Extended Data Fig. 1.
Source Data Extended Data Fig. 2
Source Data Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Source Data Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Source Data Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Source Data Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Source Data Extended Data Fig. 6.
Source Data Extended Data Fig. 7
Source Data Extended Data Fig. 7.
Source Data Extended Data Fig. 8
Source Data Extended Data Fig. 8.
Source Data Extended Data Fig. 9
Source Data Extended Data Fig. 9.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pirzgalska, R.M., Henriques-Alves, B., Raposo, B. et al. Neuroepithelial VIP–VIPR1 interactions differentially control enteric type 1 and type 2 immunity. Nat Immunol 26, 2244–2255 (2025). https://doi.org/10.1038/s41590-025-02326-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-025-02326-0