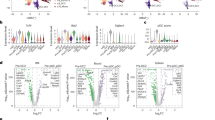
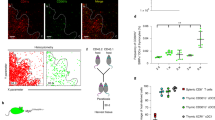
Abstract
Type 2 conventional dendritic cells (cDC2s) are functionally and phenotypically heterogenous. Previous work in mice and humans identified two cDC2 subsets (cDC2As and cDC2Bs) and a monocytic DC3 subset. However, the microenvironmental cues governing their distinct differentiation pathways remain unclear. Here, we delineate mouse cDC2 lineage relationships and the sequential signals required for cDC2A maintenance. We show that cDC2s, arising from the CLEC9A+ cDC progenitor, encompass T-BET-expressing cDC2As and two cDC2B subsets distinguished by MGL2 expression, with monocytic DC3s exhibiting transcriptional overlap with Mgl2− cDC2Bs. Among these subsets, T-BET+ cDC2As dominate the spleen where they require cell-intrinsic retinoic acid signaling to sustain their differentiation via Notch signals. Lymphotoxin-β receptor signaling on splenic cDC2s limits F-actin content retaining cDC2s at sites of retinol delivery. In summary, these data establish the developmental and transcriptional relationships between diverse cDC2 subsets and identify signals that regulate their prevalence in specific lymphoid tissues.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The mouse sequencing data are available through the Gene Expression Omnibus under accession number GSE302089. This manuscript makes use of previously published scRNA-seq data from GSE262474, GSE137710 and FigShare (https://doi.org/10.6084/m9.figshare.22232056.v1)52.
Code availability
This study did not report original code.
References
Liu, T.-T. et al. Ablation of cDC2 development by triple mutations within the ZEB2 enhancer. Nature 607, 142–148 (2022.
Tussiwand, R. et al. KLF4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928 (2015).
Lewis, K. L. et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35, 780–791 (2011).
Satpathy, A. T. et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14, 937–948 (2013).
Persson, E. K. et al. IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38, 958–969 (2013).
Brown, C. C. et al. Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell 179, 846–863 (2019).
Liu, Z. et al. Dendritic cell type 3 arises from Ly6C+ monocyte–dendritic cell progenitors. Immunity 56, 1761–1777 (2023).
Minutti, C. M. et al. Distinct ontogenetic lineages dictate cDC2 heterogeneity. Nat. Immunol. 25, 448–461 (2024).
Rodrigues, P. F. et al. Progenitors of distinct lineages shape the diversity of mature type 2 conventional dendritic cells. Immunity 57, 1567–1585 (2024).
Zhu, Y. et al. Transcription factors TCF4 and KLF4 respectively control the development of the DC2A and DC2B lineages. Nat. Immunol. 26, 1275–1286 (2025).
Briseño, C. G. et al. Notch2-dependent DC2s mediate splenic germinal center responses. Proc. Natl Acad. Sci. USA 115, 10726–10731 (2018).
Damm, K., Heyman, R. A., Umesono, K. & Evans, R. M. Functional inhibition of retinoic acid response by dominant negative retinoic acid receptor mutants. Proc. Natl Acad. Sci. USA 90, 2989–2993 (1993).
Lewis, S. M., Williams, A. & Eisenbarth, S. C. Structure and function of the immune system in the spleen. Sci. Immunol. 4, eaau6085 (2019).
Klebanoff, C. A. et al. Retinoic acid controls the homeostasis of pre-cDC-derived splenic and intestinal dendritic cells. J. Exp. Med. 210, 1961–1976 (2013).
Beijer, M. R., Kraal, G. & den Haan, J. M. Vitamin A and dendritic cell differentiation. Immunology 142, 39–45 (2014).
Dudziak, D. et al. Differential antigen processing by dendritic cell subsets in vivo. Science 315, 107–111 (2007).
Beijer, M. R. et al. A crucial role for retinoic acid in the development of Notch-dependent murine splenic CD8−CD4− and CD4+ dendritic cells. Eur. J. Immunol. 43, 1608–1616 (2013).
Zeng, R. et al. Retinoic acid regulates the development of a gut-homing precursor for intestinal dendritic cells. Mucosal Immunol. 6, 847–856 (2013).
Mora, J. R., Iwata, M. & von Andrian, U. H. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat. Rev. Immunol. 8, 685–698 (2008).
Kuwata, T. et al. Vitamin A deficiency in mice causes a systemic expansion of myeloid cells. Blood 95, 3349–3356 (2000).
Rajaii, F., Bitzer, Z. T., Xu, Q. & Sockanathan, S. Expression of the dominant negative retinoid receptor, RAR403, alters telencephalic progenitor proliferation, survival, and cell fate specification. Dev. Biol. 316, 371–382 (2008).
Kabashima, K. et al. Intrinsic lymphotoxin-β receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity 22, 439–450 (2005).
Vanderkerken, M. et al. ILC3s control splenic cDC homeostasis via lymphotoxin signaling. J. Exp. Med. 218, e20190835 (2021).
Steinhoff, J. S., Lass, A. & Schupp, M. Biological functions of RBP4 and its relevance for human diseases. Front. Physiol. 12, 659977 (2021).
Derebe, M. G. et al. Serum amyloid A is a retinol binding protein that transports retinol during bacterial infection. eLife 3, e03206 (2014).
Bang, Y. J. et al. Serum amyloid A delivers retinol to intestinal myeloid cells to promote adaptive immunity. Science 373, eabf9232 (2021).
Heng, T. S. P. et al. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Liu, D. et al. CD97 promotes spleen dendritic cell homeostasis through the mechanosensing of red blood cells. Science 375, eabi5965 (2022).
Saito, S., Watabe, S., Ozaki, H., Fusetani, N. & Karaki, H. Mycalolide B, a novel actin depolymerizing agent. J. Biol. Chem. 269, 29710–29714 (1994).
Dutertre, C. A. et al. Single-cell analysis of human mononuclear phagocytes reveals subset-defining markers and identifies circulating inflammatory dendritic cells. Immunity 51, 573–589 (2019).
Liu, Z. et al. Fate mapping via MS4A3—expression history traces monocyte–derived cells. Cell 178, 1509–1525 (2019).
Kumamoto, Y. et al. CD301b⁺ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity 39, 733–743 (2013).
Tatsumi, N., Codrington, A. L., El-Fenej, J., Phondge, V. & Kumamoto, Y. Effective CD4 T cell priming requires repertoire scanning by CD301b+ migratory cDC2 cells upon lymph node entry. Sci. Immunol. 6, eabg0336 (2021).
Liu, D., Wu, J., An, J. & Cyster, J. G. Requirements for cDC2 positioning in blood-exposed regions of the neonatal and adult spleen. J. Exp. Med. 217, e20192300 (2020).
Liu, D., Duan, L. & Cyster, J. G. Chemo- and mechanosensing by dendritic cells facilitate antigen surveillance in the spleen. Immunol. Rev. 306, 25–42 (2022).
Calabro, S. et al. Differential intrasplenic migration of dendritic cell subsets tailors adaptive immunity. Cell Rep. 16, 2472–2485 (2016).
Yokota, A. et al. GM-CSF and IL-4 synergistically trigger dendritic cells to acquire retinoic acid-producing capacity. Int. Immunol. 21, 361–377 (2009).
Guilliams, M. et al. Skin-draining lymph nodes contain dermis-derived CD103− dendritic cells that constitutively produce retinoic acid and induce Foxp3+ regulatory T cells. Blood 115, 1958–1968 (2010).
Buechler, M. B. et al. A stromal niche defined by expression of the transcription factor WT1 mediates programming and homeostasis of cavity-resident macrophages. Immunity 51, 119–130 (2019).
Levine, A. G. et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 546, 421–425 (2017).
Schraml, B. U. et al. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell 154, 843–858 (2013).
Han, H. et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14, 637–645 (2002).
Meredith, M. M. et al. Expression of the zinc finger transcription factor zDC (ZBTB46, BTBD4) defines the classical dendritic cell lineage. J. Exp. Med. 209, 1153–1165 (2012).
Wu, M. et al. Innervation of nociceptor neurons in the spleen promotes germinal center responses and humoral immunity. Cell 187, 2935–2951 (2024).
Nersesian, S. et al. Effects of modulating actin dynamics on HER2 cancer cell motility and metastasis. Sci. Rep. 8, 17243 (2018).
Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293–1308 (2018).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Heimberg, G. et al. A cell atlas foundation model for scalable search of similar human cells. Nature 638, 1085–1094 (2025).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
McInnes, L. et al. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 3(29), 861, https://doi.org/10.21105/joss.00861 (2018).
van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729 (2018).
Wang, H., Liu, Z. & Li, Z. Bulk and single-cell RNA-seq data. FigShare figshare.com/articles/dataset/Bulk_and_single-cell_RNA-seq_data/22232056/1 (2023).
Sequeira, A. M., Ijsselsteijn, M. E., Rocha, M. & de Miranda, N. PENGUIN: a rapid and efficient image preprocessing tool for multiplexed spatial proteomics. Comput. Struct. Biotechnol. J. 23, 3920–3928 (2024).
Greenwald, N. F. et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 40, 555–565 (2022).
Windhager, J. et al. An end-to-end workflow for multiplexed image processing and analysis. Nat. Protoc. 18, 3565–3613 (2023).
Acknowledgements
We thank C. Ware for agonist antibodies to LTβR (rat IgG anti-mouse LTβR clone 4H8), Sanford Burnham Prebys Institute, the veterinary staff at the Division of Comparative Medicine and the staff at the Faculty of Medicine Flow Cytometry Core Facility (University of Toronto) for support and all members of the laboratory of J.L.G. for help. We are grateful for funding support from the Canadian Institutes for Health Research/Instituts de Recherche en Santé du Canada (CIHR/IRSC) to J.L.G. (FDN-159922 and PJT-195705) and FRN-165973 to C.J.G. Flow cytometry was performed at the Centre for Immune Analytics Flow Cytometry Facility, University of Toronto, and the SickKids Flow Cytometry Facility, supported by the SickKids Foundation and a CFI John Evans Fund Leaders grant to C.J.G. This work was supported by a Parker Institute for Cancer Immunotherapy Senior Fellowship (C.C.B.), an NIH NIAID DP2 award (DP2AI171116; C.C.B.), Pew Biomedical Scholar Award (C.C.B.) and Josie Robertson Investigator Award (C.C.B.). A.A.N. was supported by a CIHR Frederick Banting and Charles Best Canada Graduate Scholarship Doctoral (CGS-D) Award (FRN-165746). L.F. is supported by an MSK Houghton-Coit Fellowship. We acknowledge the use of the Integrated Genomics Operation Core, MSKCC, funded by the NCI Cancer Center Support Grant (CCSG, P30 CA08748), Cycle for Survival and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. J.L.G receives salary support as a Canada Research Chair in tissue specific immunity.
Author information
Authors and Affiliations
Contributions
A.A.N., L.F., J.L.G. and C.C.B. designed, conceptualized and conducted the experiments, analyzed the data and wrote the manuscript. A.A.N. conducted the experiments, with assistance from J.S.Y.A., D.C.D., C.L., L.A.W., A.A.W. and B.M. M.Z. and T.M. performed the imaging analyses. T.P., A.Y. and Z.T. performed scRNA-seq data analyses. K.H. aided in data analysis and presentation. M.S.G., C.J.G. and C.L.M. provided mutant mice and mouse BM. D.T. and J.S.A. provided Myc. B reagent and contributed to the design of the F-actin inhibition experiments in mice. J.L.G. and C.C.B. supervised the project. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Carlos Minutti and the other anonymous reviewer for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: L. A. Dempsey, in collaboration with the rest of the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 cDC2 development and heterogeneity.
a, Gating strategy for identification of cDC2 subsets in skin-draining peripheral lymph nodes (pLN) of Clec9aCre/CreTbx21RFP-CreERT2R26lsl-YFP mice. b, Representative flow cytometry of T-BET− cDC2 from indicated lymph nodes of Clec9aCre/CreTbx21RFP-CreERT2R26lsl-YFP mice showing expression of CD16/32 and summary graph showing frequency of CD16/32+ cells among Clec9a-fate mapped (YFP+) and non-fate mapped CD11b+T-BET− DCs (n = 4 mice). c, Gating strategy for FACS isolation of YFP+ and YFP− Lin−MHCII+ cells from spleen and lymph nodes of Clec9aCre/CreTbx21RFP-CreERT2R26lsl-YFP mice for scRNA-seq analysis. d, Representative flow cytometry of splenic cDC2s showing expression of MGL2 and CLEC10A (n = 3 mice). e, Violin plot depicting the expression of Zbtb46 in YFP+ and YFP− cells across clusters defined in Fig. 1b. f, Uniform manifold approximation and projection (UMAP) visualization of dendritic cells profiled by scRNA-seq in Rodrigues et al., colored by cluster. g, UMAP overlaid by expression of indicated genes. h, Heatmap showing scaled, imputed expression of all DEGs (pairwise comparison, fold change>1.5, adjusted P < 0.01) for indicated cDC2A, cDC2B and DC3 clusters. i, Violin plots depicting the expression of tdTomato in indicated clusters (as in f) derived from Cd300cCre-hCD2R26tdTomato, Il7rCreR26tdTomato, and Ms4a3CreR26tdTomato mice. j, Schematic showing cDC2 development and heterogeneity with transcriptional overlap between subsets indicated by dashed box. Data in in a, b, and d are representative of 2 independent experiments. Data are mean ± s.e.m., each symbol represents an individual mouse. Unpaired two-tailed t test; *P < 0.05, **P < 0.01.
Extended Data Fig. 2 Vitamin A and RAR signaling are required for optimal generation of cDC2 numbers.
a-c, Compiled data showing (a) cDC2/cDC1 ratio (b) %ESAM expression and (c) MFI of ESAM on cDC2 in mice given a VAD or control diet. DCs were distinguished identified based on Live B220−CD11c+MHCIIhi singlets, before being delineated into CD8+ cDC1 or CD11b+ cDC2 (n = 8 per group). d, Representative IMC images of three different 7 μm spleen cross-sections (10X magnification) depicting DCIR2+ cDC2 (red) within marginal zone bridging channels (MZBCs) in control and VAD diet animals. CD169 (blue pseudocolour) was used to outline the marginal zone; B220 (green pseudocolour) and CD3 (red pseudocolour) were used to outline the B cell follicle. Average DCIR2 pixel intensity was measured in MZBCs of control and VAD diet animals (n = 5 per group). e, Representative images of three different 7 μm spleen cross-sections (10X magnification) depicting spleen follicles using CD3 (red pseudocolour), CD169 (blue pseudocolour), and B220 (green pseudocolour). (Bottom) Quantification of average follicle size across control and VAD diet animals. Scale bars indicate 250μm (n = 5 per group). f-h, Compiled data showing (f) cDC2/cDC1 ratio (g) %ESAM expression and (h) MFI of ESAM on cDC2 in mice supplemented with RA or vehicle control for 9 days (n = 7 per group for f and g, n = 4 per group for h). i, Compiled data showing cDC1 frequencies in mice given a Control Diet (n = 6) or VAD diet (n = 5). j, Compiled data showing cDC1 frequencies in mice supplemented DMSO control or RA (n = 5 per group). k, Compiled data showing absolute numbers of CD11b+ cDC2 and CD8+ cDC1 in the PPs of VAD vs control diet animals (n = 5 per group). Data in a-g, i-k are representative of two independent experiments, and h is representative of one experiment. Statistics were measured by Mann-Whitney U test in a, b, c, d, e, f, g, h, i, and j (ns, P > 0.05, *P < 0.05, **P < 0.01, and ***P < 0.001) and multiple Mann-Whitney tests with Holm Sidak’s adjustment in k (ns, P > 0.05 and ***P < 0.001). Error bars represent the mean ± s.d., each symbol represents an individual mouse.
Extended Data Fig. 3 Characterization of Tbx21 expression in cDC subsets.
a, Representative plot showing ESAM and RFP (T-BET) expression in Sirpa+ cDC2 in the spleen (left) and Peyer’s Patches (right) of Tbx21RFP-Cre mice. b, Histogram showing ESAM expression is enriched in splenic cDC2A compared to cDC2B. c, Flow cytometry gating strategy to identify cDC2A. In brief, cDC were distinguished from other immune cell populations in the spleen, B220+, Ly6Cint/hi, and Tbx21hi cells (top-left to top-right) were gated out to remove B cells, plasmacytoid DCs, monocytes, neutrophils, and contaminating NK cells and T cell populations (in contrast, cDCs are express moderate levels of Tbx21). Next, the cDC population was gated on by their high surface expression of CD11c and MHCII. Expression of Sirpa and XCR1 was used to distinguish cDC2 vs cDC1 populations respectively (bottom-left and bottom-center). Note that Tbx21 expression is only observed in the cDC2 and not in cDC1. B6 mice lacking the RFP reporter for Tbx21 expression were used as a negative control (bottom-right).
Extended Data Fig. 4 Cellularity and immune cell composition in VAD spleens.
a-g, absolute numbers of (a) total live CD45+ cells and (b) B cell, (c) T cell, (d) macrophage, (e) neutrophil, (f) monocyte, and (g) NK cell populations in the spleen were compared between 10-12-week-old mice fed a Vitamin A-sufficient control or VAD diet since embryonic day 14 (n = 6 per group). Data in a-g are representative of two independent experiments. Mann-Whitney U test was conducted for comparison between two groups in a, b, d, and e (ns, P > 0.05 and **, P < 0.01). 2-Way ANOVA with Tukey’s multiple comparisons test was used for comparison between multiple groups across two categorical variables in c, f, and g (ns, P > 0.05, *P < 0.05, and ***P < 0.001). Error bars represent the mean ± s.d., each symbol represents an individual mouse.
Extended Data Fig. 5 Impact of cDC-intrinsic dnRAR expression on pre-cDC2A and cDC2A in different tissues.
a, Representative flow cytometry plots showing gating strategy for the identification of pre-cDC1, pre-cDC2A, and pre-cDC2B subsets in the BM. b, Compiled data of pre-cDC subsets in the BM of littermate controls (n = 8) vs Zbtb46CrednRaralsl/lsl (n = 9). c, Representative flow cytometry plots comparing cDC2A populations between the spleen and pLN of Tbx21RFP-CrednRaralsl/lsl and Tbx21RFP-Cre littermate controls. d, Compiled data showing frequency of T-BET expression in pLN cDC2s from Tbx21RFP-Cre littermate controls and Tbx21RFP-CrednRaralsl/lsl (n = 6 per group). e, Schematic of mixed competitive BM chimera approach to study cDC2A-intrinsic loss of RAR signaling in vivo. f, Representative flow cytometry plots showing ESAM and T-BET RFP expression from CD45.1+ B6 and CD45.2+ Tbx21RFP-Cre or Tbx21RFP-CrednRaralsl/lsl donors in chimeras from (e). g-i, Compiled data of f showing ESAM expression in CD45.2+ cDC2 containing Tbx21RFP-Cre or Tbx21RFP-CrednRaralsl/lsl donor-derived cells (g), and CD45.2/CD45.1 chimerism of ESAM+ cDC2A (h) and ESAM− cDC2B (i) (n = 5 per group). Data in b, d, and g-i are representative of two independent experiments. in a and b. Statistics were measured by Mann-Whitney U test in b, d, g, h, and i (ns, P > 0.05, **P < 0.01). Error bars represent the mean ± s.d., each symbol represents an individual mouse.
Extended Data Fig. 6 Optimal generation of splenic cDC2A requires cDC-intrinsic LTbR signaling prior to expression of Tbx21.
a, Relative expression of Ltbr mRNA transcripts measured by qPCR in spleen cDC subsets in (a) control diet mice (n = 12) vs VAD diet-fed mice (n = 9). b, Relative expression of Ltbr mRNA transcripts measured by qPCR in spleen cDC2 of mice given a single dose of DMSO, RA after 2- or 24-hours (n = 5 per group). c, d, Surface expression of LTβR measured using anti-LTβR (3C8) antibody in (c) cDC1 or (d) cDC2 of mice given a single dose of DMSO (n = 8), RA after 2 (n = 9) or 24 h (n = 10). e, Representative histograms showing LTβR surface expression (clone 3C8) in splenic cDC2A, cDC2B, and cDC1 populations (as indicated in figure legend (far-right)) in C57Bl/6 (left), Tbx21RFP-Cre (center), and Tbx21RFP-CreLtbrfl/fl (right) mice. f, Compiled data of e showing LTβR surface expression between Tbx21RFP-CreLtbrfl/fl compared to littermate controls (n = 4 per group). g, Representative histograms depicting ESAM expression in cDC2s of Zbtb46CreLtbrfl/fl vs. littermate controls. h, Compiled data of g showing % ESAM expression on splenic cDC2 in littermate controls (n = 4) vs Zbtb46CreLtbrfl/fl mice (n = 7). Data in a, c, d, and h are representative of two independent experiments; b and f are representative of one experiment. Statistics were measured by multiple Mann-Whitney tests with Holm Sidak’s adjustment in a (ns, P > 0.05), Kruskal-Wallis test with Dunn’s multiple-comparisons test in b, c, and d (ns, P > 0.05 and *P < 0.05), and Mann-Whitney U tests in f and h (*P < −0.05). Error bars represent the mean ± s.d., each symbol represents an individual mouse.
Extended Data Fig. 7 RBPR2 (Stra6l) gene/protein expression is enriched in cDC2A.
a, UMAP plot depicting Stra6l gene expression in cDC populations from Fig. 1 scRNA-seq dataset of skin-draining pLN, mLN, and medLN dataset, with accompanying legend of DC clusters.
Extended Data Fig. 8 Phalloidin staining index of splenic cDC2A ex vivo and during Myc. B treatment conditions.
a, Representative flow plots and histograms showing Phalloidin staining in pLN stromal cell subsets (top-left and top-right: CD31+ ECs (blue) vs, CD31+Pdpn+ LECs (red) vs. CD31−Pdpn+ FRCs (orange)) and splenic cDC2 subsets (bottom-left and bottom-right: –FMO staining control (blue) compared to T-BET+ cDC2A (red) and T-BET− cDC2B (orange)). b, Representative plots showing Phalloidin staining of ex-vivo sorted splenic cDC2 after culture in Myc. B at labeled concentrations (or DMSO vehicle control) at 37 °C for 2 h and compiled MFI data on gated T-BET+ cDC2A (right). c, Compiled data of b. Each point on plot is representative of three technical replicates from two pooled Tbx21RFP-Cre spleens.
Supplementary information
Source data
Source Data for Figs. 1–6 and Extended Data Figs. 1, 2, 4–6 and 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nguyen, A.A., Fisher, L., Ahn, J.S.Y. et al. Sequential lymphotoxin-β receptor and retinoic acid receptor signals regulate cDC2 fate. Nat Immunol 26, 2159–2169 (2025). https://doi.org/10.1038/s41590-025-02329-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-025-02329-x