Abstract
We present ReDU (https://redu.ucsd.edu/), a system for metadata capture of public mass spectrometry-based metabolomics data, with validated controlled vocabularies. Systematic capture of knowledge enables the reanalysis of public data and/or co-analysis of one’s own data. ReDU enables multiple types of analyses, including finding chemicals and associated metadata, comparing the shared and different chemicals between groups of samples, and metadata-filtered, repository-scale molecular networking.
Similar content being viewed by others
Main
The availability of public mass spectrometry (MS)-based metabolomics data continues to grow, but leveraging these data has been difficult. It is arduous to find relevant files scattered among different datasets and analyze them in a consistent and meaningful manner. Therefore, we developed the Reanalysis of Data User (ReDU) interface (https://redu.ucsd.edu/), a community-minded approach that addresses these challenges. ReDU is a repository-scale analysis system using consistent formatting and controlled vocabularies that can be validated. ReDU finds uniformly formatted public MS/MS data in the Global Natural Product Social Molecular Networking Platform (GNPS; https://gnps.ucsd.edu/) via formatted metadata1. New or previously collected data can be added, provided they adhere to the ReDU metadata standards (the implemented drag-and-drop validator is applicable to any scientific data) and the data are available in GNPS-MassIVE repository. Further, ReDU has built-in analyses and can launch co- or reanalysis of data via GNPS; it enables reanalysis of MS/MS data de novo as opposed to the meta-analysis of reported results.
Simple but important questions can be explored using repository-scale public data. For example, of those sampled, what human biospecimen or sampling location is best for detecting a given drug? What molecules have been observed in humans <2 years old? Current metabolomics repositories (for example, GNPS/MassIVE, MetaboLights2, Metabolomics Workbench3) contain data and metadata; however, finding individual files typically requires manual navigation, conversion of different file formats and reformatting of inconsistent metadata formats.
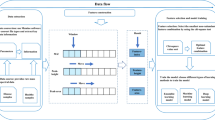
ReDU enables users to find and choose files (Fig. 1a) via consistent and validated sample information (that is, metadata) created by users with a template. The template uses controlled vocabularies and ontologies (for example, NCBI Taxonomy4, UBERON5, DOID6 and MS ontology). ReDU automatically incorporates public data into the GNPS/MassIVE repository with the corresponding ReDU-compliant metadata file. Currently, 38,305 files in GNPS (19.6% of GNPS) are ReDU compatible. These include data collected from natural and human-built environments, human and animal tissues, biofluids and food together with other data from around the world (Extended Data Fig. 1), which were analyzed using different instruments, ionization methods, sample preparation methods, etc. From the 103,230,404 MS/MS spectra included in ReDU, 4,528,624 spectra were annotated (rate of 4.39%, ~1% false discovery rate (FDR)) as one of 13,217 unique MS/MS library matches (level 2 or 3) (Supplementary Table 1; refs. 7,8).
a, ReDU provides users the tools to find public data in the GNPS/MassIVE knowledgebase and explore public data analyses in ReDU, and it enables repository-scale co- and reanalyses in GNPS. Contributors are provided a template for sample information and a drag-and-drop validator. b, Two-dimensional Emperor plot displaying the projection of human plasma samples, n = 31 (orange) from patients with rheumatoid arthritis (not included in ReDU), onto files (points) in ReDU, n = 34,003 (colored by UBERON ontology) (NCBI Taxonomy-based opacity used: projected data, 1.0; 9606|Homo sapiens, 0.7; all other data, 0.25). c, Illustrative results from Chemical Explorer for 12-ketodeoxycholic acid, cholic acid and rosuvastatin annotated in human fecal (n = 5,097) files over different life stages. d, Group Comparator performed on human blood (n = 711), fecal (n = 5,097) and urine (n = 307) samples resulted in different chemical compositions as illustrated by bilirubin, urobilin and stercobilin.
The uniformity of information in ReDU enables metadata-based and repository-scale analyses, including repository-scale principal-component analysis (PCA) based on the annotations of each file. In Fig. 1b, the chemical similarity of files in ReDU, based on MS/MS annotations, is plotted in Emperor9, an interactive visualization tool, onto which new samples can be projected using a GNPS taskID. ReDU also includes a tool called Chemical Explorer, which enables selection of a molecule and retrieval of its associations with the metadata, also known as sample information association. For instance, querying 12-ketodeoxycholic acid (filtering to include human feces) revealed that it was observed after infancy (Fig. 1c), whereas cholic acid displayed the opposite trend. This observation is attributed to the developing gut microbiome, which converts primary bile acids into secondary bile acids, and suggests that early in life the microbes that do such conversions are not present10,11. Similarly, rosuvastatin, a lipid-lowering drug, was found in adults, matching prescription demographics12.
The Group Comparator tool compares user-selected groups (selected with metadata) and tabulates the annotation information, and subsequent user interpretation can determine which chemicals are similar or different between groups, such as human blood, feces and urine (Fig. 1d) or Staphylococcus aureus, Bacillus subtilis and Streptomyces cultures (Extended Data Fig. 2). Group Comparator analysis of 6,115 human blood, fecal and urine samples indicated differences in the percentage of files in which bile pigments were observed. Bilirubin was more frequently annotated in blood, and urobilin and stercobilin were most often annotated in feces. Similarly, comparison of MS data from bacterial cultures revealed differences in annotation of pyroglutamylisoleucyllysine (PyroGlu-Ile), staurosporine and surfactin-C14. While the rationale for the increased percentage of PyroGlu-Ile in S. aureus is unknown, staurosporine is a known secondary metabolite produced by Streptomyces13 and surfactin-C14 is a known secondary metabolite produced by B. subtilis14.
ReDU can be used to select files using metadata and launch repository-scale molecular networking. Figure 2a displays the result of repository-scale selection and molecular networking (results with MolNetEnhancer are shown in Extended Data Fig. 3; ref. 15) of human blood, urine and fecal samples. In total, 6,663 nodes in the molecular network (created from 399,826 MS/MS spectra) were annotated (Fig. 2b) via spectral library matching (level 2 or 3; ref. 8). While the annotation percentage was relatively low (7.58% of nodes), molecular networking linked chemicals with similar MS/MS patterns. As MS/MS patterns are often coupled to chemical structure, one can propagate annotations via analogy in combination with mass differences, exact mass and manual interpretation of the MS/MS spectra. Simply put, repository-scale molecular networking improves the ability to annotate unknown chemical analogs across different datasets or sample types. For example, we propose clindamycin analogs (compounds 2–9) through propagation (for example, on the basis of changes in m/z ratio and MS/MS spectral interpretation), some of which match reported metabolites such as clindamycin sulfoxide (compound 4; ref. 16), from the annotation of clindamycin (compound 1). The clindamycin analogs (compounds 2–9) were linked to clindamycin (compound 1) across human urine, blood and fecal data originating from different datasets (Fig. 2c, Supplementary Discussion and Supplementary Figs. 1–4).
a, Molecular network of human blood (n = 711), fecal (n = 5,097) and urine (n = 307) samples in ReDU with nodes colored by annotation status (red, annotated; gray, unannotated). b, A summary of MS/MS library matching results (level 2 or 3) is displayed for the nodes in the network and all MS/MS spectra considered in the molecular network. c, A component of the repository-scale molecular networking containing clindamycin. Nodes are colored by the sample type. Node size reflects the number of MassIVE datasets. Node shape represents annotation status (diamond, annotated; circle, unannotated). Putatively annotated clindamycin analogs (compounds 2–9), based on MS/MS interpretation, are indicated using dark blue dashed arrows and numbers, corresponding to the proposed structures.
Lastly, all data in ReDU, including the metadata and annotation information, are available for download from the homepage. The annotation information was used for molecular cartography17 at the repository scale, which was used to plot the location of drugs in human samples (Extended Data Fig. 4 and Supplementary Video 1). We envision that this information will be invaluable to researchers. ReDU’s utility will continue to grow as more data are uploaded to GNPS/MassIVE and as public MS/MS reference libraries expand, scaling in breadth and depth. ReDU is a resource developed for the community and strives to embody the findable, accessible, interoperable, and reusable (FAIR) principles18.
Methods
ReDU content
The homepage of ReDU (https://redu.ucsd.edu/) is the launch point for different analyses, centered around ‘Analyze Your Data’ or ‘Analyze Public Data’. It also links to ‘Documentation’, ‘How to Contribute Data’, ‘ReDU Sample Information Validator’, ‘Download Database’ and ‘File Query—Sample Information’. The ‘Documentation’ option (Supplementary Fig. 5a) links to the ReDU documentation, and the ‘How to Contribute Data’ option (Supplementary Fig. 5b) links to the subsection of documentation that lists the steps necessary to contribute data to ReDU. The ‘ReDU Sample Information Validator’ (Supplementary Fig. 5c) links to a drag-and-drop validator (https://redu.ucsd.edu/ReDUValidator) that verifies that the sample information template required for data contribution adheres to the required formatting and terms in a controlled vocabulary (additional terms must be submitted via GitHub at https://github.com/mwang87/ReDU-MS2-GNPS/issues). Supplementary Fig. 5d links to a text field into which a file name can be queried and any associated metadata are displayed. ‘Download Database’ (Supplementary Fig. 5e) downloads all the sample information included in ReDU in a tab-separated text file. ‘Download Annotations’ (Supplementary Fig. 5f) downloads all the MS/MS annotations. Links to specific analyses are detailed below. The ReDU server is built using the Python flask framework, SQLite and a Vue.js front end.
Data and sample information contribution
Data files (.mzXML or .mzML) and a ReDU-validated sample information (metadata) table are necessary for inclusion of data in ReDU and must be uploaded to a public MassIVE dataset. A sample information template and validator are provided. Detailed step-by-step instructions can be found in the ReDU documentation (https://mwang87.github.io/ReDU-MS2-Documentation/HowtoContribute/).
Chemical annotations based on MS/MS reference library matches
MS/MS data were reanalyzed in a consistent manner to provide chemical annotations based on spectral library matches. The search was performed on the MS/MS product ion scans in files located in MassIVE de novo (that is, original MS/MS data and not the reported results) using GNPS’ default parameters. The resulting MS/MS spectral matches (that is, annotations) were counted per file and tabulated; multiple hits to the same CCMSLIB ID in the same file were counted once. All annotation information was downloaded from ReDU (Supplementary Fig. 5f) and processed in R. Script is available on GitHub in the examples folder (https://github.com/mwang87/ReDU-MS2-GNPS/tree/master/examples). Supplementary Table 1 displays the number of MS/MS reference library spectra available in each library in GNPS (for example, GNPS-LIBRARY) and the total number of annotations in ReDU per library. Further information can be found at https://proteomics2.ucsd.edu/ProteoSAFe/result.jsp?task=ba6a5b6a1c0946b3a641c67ad59fb2df&view=production_library_sizes#%7B%22table_sort_history%22%3A%22main.number_spectra_dsc%22%7D.
Principal-component analysis
PCA was performed on the counts of each chemical annotation from GNPS spectral library matching using GNPS’ default parameters (https://mwang87.github.io/ReDU-MS2-Documentation/). PCA was performed in Python with scikit-learn. The eigenvector matrix was retained and used to calculate the location of the projected points.
Multivariate analysis of public data
Emperor (https://github.com/biocore/emperor) was used to generate interactive visualizations using the results from PCA (Supplementary Fig. 5g). Emperor has many plotting options (including the axes and the color of points based on sample information) and filtering options and can rescale data. Clicking on any of the points in the plot causes the file name to be displayed in the bottom-left corner. The plot can be saved as an image file. Additional instructions on Emperor can be found in its online documentation (http://emperor.microbio.me/uno/).
Comparing user data to public data via multivariate analysis
Users can co-analyze their data via projection onto an Emperor plot of all data in ReDU (Supplementary Fig. 5h and Fig. 1b). Users submit their data by providing a GNPS taskID into the field. GNPS library search, GNPS molecular networking and GNPS feature-based molecular networking are compatible. It is encouraged that default library search parameters be used. The taskID provides the information required to calculate the coordinates for the projection of samples onto the precalculated PCA plot (visualized using Emperor) of all ReDU data. Projection was performed by multiplying the annotations for each file (vector) by the eigenvectors to calculate the location of data points in the precalculated coordinate frame. The user can highlight their data using the ‘Your Data’ term in the ‘type’ category; we suggest using this column to change the scale or opacity of the sample points to visualize user data.
In the example shown in Fig. 1b, human plasma samples not yet entered in ReDU at the time of data analysis were subjected to a GNPS library search using default parameters; the data and illustrative library search can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f39c94cb7afe4568950bf61cdb8fee0d. The taskID was entered using the ‘Compare Your Data to Public Data via Multivariate Analysis’ option (https://mwang87.github.io/ReDU-MS2-Documentation/), resulting in the Emperor plot. The example button populates the field with the taskID used to generate the figure. The following settings were used to create the image. Points were scaled using the UBERONBodyPartName category and globally scaled to 1.3 with the exception of blood, blood plasma and blood serum, which were scaled to 2.5, and the projected data were scaled to 5 (nan). The opacity was set to 0.25 globally, and, using the NCBITaxonomy column, the values for the projected data were set to 1 (nan) and for all 9606|Homo sapiens data were set to 0.7. Points were colored based on UBERONBodyPartName. All points were set to gray (#d1d1d1), except skin samples (blue, #91bfdb), blood samples (red, #d73027), feces (purple, #998ec3) and the projected data (orange, #f1a340). A .json file (settings file) has been provided at GitHub (https://github.com/mwang87/ReDU-MS2-GNPS/tree/master/examples) to reproduce the plot by uploading it in the ‘load saved settings’ option. This example is only intended to illustrate that blood samples cluster closely with other blood samples already in the ReDU database. Note that periodic updates to the ReDU database will shift the appearance of the data over time. The code and materials needed to recreate this analysis and plots are available on GitHub at https://github.com/mwang87/ReDU-MS2-GNPS/tree/master/examples.
Chemical Explorer
The Chemical Explorer can be accessed on the ReDU homepage (Supplementary Fig. 5i). The chemical annotations resulting from library search, described above, were used to populate the Chemical Explorer table (https://mwang87.github.io/ReDU-MS2-Documentation/). A search box is provided for queries. Note that the chemical name that appears reflects that which is entered in the spectral references databases (Supplementary Table 2) included in GNPS and is case sensitive. The sample information associated with a particular chemical can be accessed by clicking the ‘View Association’ button, as well as a list of files in which the chemical was found by clicking the ‘View Files’ button. The sample information is tabulated for the selected chemical and ranked based on the proportion of files associated with a sample information term. The Chemical Explorer can also be used on a subset of data, selected using the ReDU file selector (Supplementary Fig. 5j,k) and launched by hitting the ‘Launch Chemical Explorer’ button under the ‘Analyze Public Data’ section. Note that only files placed into group 1 (G1) are considered in the calculation of the associated sample information.
In the example shown in Fig. 1d, the file selector was used to filter only human files (NCBItaxonomy = 9606|Homo sapiens), fecal samples were filtered using UBERONBodyPartName and samples were selected into G1 based on Lifestage (samples marked as not applicable, not collected or not specified were excluded). Chemical Explorer was launched. The resulting webpage was searched using the search box for illustrative examples, specifically ‘Spectral Match to 12-Ketodeoxycholic acid from NIST14’, ‘Cholic acid’ and ‘Stercobilin’. The ‘View Associations’ button was clicked for each. The table can be downloaded using the ‘Download’ button. In this manuscript, the resulting table displayed on the ReDU website was copied and pasted into Excel (Microsoft). All associations were tabulated in a single spreadsheet, and an additional column indicating the chemical was added. The data file was saved as a tab-delimited text file and imported into R for plotting. The x axis corresponds to the following life stages: infancy (<2 years), n = 1,859; early childhood (2 years < x ≤ 8 years), n = 93; adolescence (8 years < x ≤ 18 years), n = 169; early adulthood (18 years < x ≤ 45 years), n = 995; middle adulthood (45 years < x ≤ 65 years), n = 933; and later adulthood (>65 years), n = 325. The code and materials needed to recreate this analysis and plots are available on GitHub (https://github.com/mwang87/ReDU-MS2-GNPS/tree/master/examples).
Group Comparator
Users can compare the occurrence of chemical annotations between two or more groups populated in the file selector by clicking the ‘Launch Group Comparator’ button after data selection (https://mwang87.github.io/ReDU-MS2-Documentation/) in the ReDU file selector (Supplementary Fig. 5j,k). GNPS chemical annotations are tabulated with the number of files in which they are found (and the percentage of files) in each group (G1–G6). This information is precalculated from library search (same information used for PCA and Chemical Explorer) using default library search parameters.
In the example shown in Fig. 1d, the file selector was used to filter only human files (NCBItaxonomy = 9606|Homo sapiens). Blood plasma (n = 678) and blood serum (n = 33) files were selected into G1 (considered together as blood), fecal (n = 5,097) files were selected into G2 and urine files (n = 307) were selected into G3. Group Comparator produced a tabulation of chemicals and corresponding counts (that is, number of times annotated) in each group. The table (.csv) was downloaded using the ‘Download’ button. The data file was imported into R for plotting. The code and materials needed to recreate this analysis and plots are available on GitHub (https://github.com/mwang87/ReDU-MS2-GNPS/tree/master/examples).
In the example shown in Extended Data Fig. 2, the file selector was used to filter only bacterial cultures (SampleType = culture_bacterial). 1423|Bacillus subtilis (n = 89) files were selected into G1, 1280|Staphylococcus aureus (n = 49) files were selected into G2 and 1883|Streptomyces (n = 7) files were selected into G3. The NCBITaxonomy metadata category was used for file selection. Group Comparator was launched. Surfactin-C14 (IUPAC ID: 3-[(3R,6S,9R,12R,15S,18R,21R,25S)-9-(carboxymethyl)-25-(9-methyldecyl)-3,6,15,18-tetrakis(2-methylpropyl)-2,5,8,11,14,17,20,23-octaoxo-12-propan-2-yl-1-oxa-4,7,10,13,16,19,22-heptazacyclopentacos-21-yl]propanoic acid; CCMS identifier: CCMSLIB00000478649), PyroGlu-Ile and staurosporine were plotted as examples. The table (.csv) was downloaded using the ‘Download’ button and imported into R for plotting. The code and materials needed to recreate this analysis and plots are available on GitHub (https://github.com/mwang87/ReDU-MS2-GNPS/tree/master/examples).
Repository-scale molecular networking and library search
Users can reanalyze with public data by clicking the ‘Reanalyze Public Data at GNPS’ text (Supplementary Fig. 5j), which links to the ReDU file selector (https://mwang87.github.io/ReDU-MS2-Documentation/). The ReDU file selector allows one to select (and filter) files based on the sample information and place multiple types of files into one of six different groups (G1–G6) for molecular networking via GNPS. Library search without molecular networking, providing annotations only, can be formed via GNPS; however, all files should be placed in G1, as groups are not supported. Upon completion of data selection, the user can launch the ‘Reanalyze with GNPS Molecular Networking’ or ‘Reanalyze with GNPS library search’ buttons, which populate the GNPS molecular networking or GNPS library search launch pages, respectively. The suggested parameters for molecular networking and library search are detailed in the GNPS documentation (https://ccms-ucsd.github.io/GNPSDocumentation/). A maximum of 5,000 files for molecular networking is suggested. Note that a free account on GNPS is required and the user must be logged in before attempting to launch reanalyses in GNPS.
In the example shown in Fig. 2, molecular networking was performed in GNPS after selecting human blood plasma and serum (n = 711), human urine (n = 307) and human fecal (n = 5,097) files in the ReDU file selector (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=a75aa494e927481dae6de12608e5e4a0). The data were filtered by removing all MS/MS peaks with m/z values ±17 with respect to the precursor’s m/z. MS/MS spectra were window filtered by choosing only the top six peaks in windows located ±50 m/z with respect to each peak throughout the spectrum. The data were then clustered with MS-Cluster with a precursor m/z tolerance of 0.02 and an MS/MS fragment ion (that is, product ion) m/z tolerance of 0.02 to create consensus spectra. Further, consensus spectra that contained fewer than five spectra were discarded. A network was then created where edges were filtered to have a cosine score above 0.7 and more than five matched peaks. Further edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top ten most similar nodes. The spectra in the network were then searched against GNPS’ spectral libraries. The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least five matched peaks. The network was opened in Cytoscape (3.7.1; https://cytoscape.org/)19, and the networks were output as a .pdf and assembled in Adobe Illustrator. The molecular networking component associated with clindamycin was analyzed using the in-browser network visualization at https://gnps.ucsd.edu/ProteoSAFe/result.jsp?view=network_displayer&componentindex=2892&task=a75aa494e927481dae6de12608e5e4a0#%7B%7D. Universal spectrum identifiers were generated (Supplementary Table 1) and used to plot the spectra displayed in Supplementary Figs. 2 and 4. MolNetEnhancer4 was launched from the results page of the molecular networking job (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=5ce4c3be9f5a4adfa1c50c9e99c4aeaf; Extended Data Fig. 3). Upon completion, the molecular network was downloaded and opened in Cytoscape. The code and materials needed to recreate this analysis and plots are available on GitHub (https://github.com/mwang87/ReDU-MS2-GNPS/tree/master/examples).
Co-analysis of user data with public data using molecular networking
Users can co-analyze their data with public data by clicking the ‘Co-analyze Your Data with Public Data at GNPS’ text, which links to the ReDU file selector (Supplementary Fig. 5k). Once the user has selected the public files they wish to include, a click of the ‘Co-analyze with GNPS Molecular Networking’ or ‘Co-analyze with GNPS Library Search’ button will load the public files into a GNPS molecular networking or GNPS library search launch page, respectively, at which point the user can add their own files to the appropriate group and submit the job. Details on molecular networking and library search can be found in the GNPS documentation (https://ccms-ucsd.github.io/GNPSDocumentation/). A maximum of 5,000 files for molecular networking is suggested. Note that a free account on GNPS is required and the user must be logged in before attempting to launch reanalyses in GNPS. If more than 5,000 files are to be co-networked, then we suggest contacting the authors, as more computing resources will need to be allocated.
Illustrative use of the ReDU database: molecular cartography
In the example shown in Extended Data Fig. 1, the ReDU information (MSV000084206) was downloaded and the latitudinal and longitudinal data were cleaned of any non-adherent formatting. The number of unique files associated with each latitude and longitude coordinate was calculated as well as the number of chemical annotations. The sum of the chemical annotations per latitude and longitude coordinate was divided by the number of unique files associated with the coordinates. Files lacking coordinates were removed. The values were log10 scaled to aid in visualization. The data were plotted in R (‘ggmap’ and ‘map’ packages were used) onto a world map. The code and materials needed to recreate this analysis and plots are available on GitHub (https://github.com/mwang87/ReDU-MS2-GNPS/tree/master/examples).
In the example shown in Extended Data Fig. 4, the ReDU information (MSV000084206) was downloaded and merged with the sample information database. A list of curated tags was generated from the curated source information table (provided). The files associated with humans were included and the chemical annotations associated with drugs or drug metabolites, putatively, were included. The number of chemical annotations per UBERON body part was divided by the number of files included for each body part. An image of an androgynous human was created in Illustrator (Adobe) and saved as a .png. The pixel coordinates associated with each label were tabulated by UBERON ontology name and merged with the ReDU drug table. The resulting file was exported as a .csv file for use in ‘ili. Files and a .json file that can be used to reproduce the illustrative example in the manuscript are available on GitHub (https://github.com/mwang87/ReDU-MS2-GNPS/tree/master/examples). The results were compiled into a video (Supplementary Video 1; https://www.youtube.com/watch?v=dzAqjBNmqPU&feature=youtu.be).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Source data for the results presented in this paper are available on GitHub (https://github.com/mwang87/ReDU-MS2-GNPS/tree/master/examples). All curated sample information can be downloaded from the ReDU homepage (https://redu.ucsd.edu/) by selecting ‘Download Database’. The current version of the ReDU information used to generate the results in this paper is archived in the GNPS/MassIVE repository (http://gnps.ucsd.edu). The accession number is MSV000084206 (https://doi.org/10.25345/C5407D).
Code availability
All software is citable using https://doi.org/10.5281/zenodo.3924422. Up-to-date developments of ReDU are available in GitHub (https://github.com/mwang87/ReDU-MS2-GNPS), with corresponding documentation (https://github.com/mwang87/ReDU-MS2-Documentation).
References
Wang, M. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016).
Haug, K. et al. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 48, D440–D444 (2020).
Sud, M. et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–D470 (2016).
Federhen, S. The NCBI Taxonomy database. Nucleic Acids Res. 40, D136–D143 (2012).
Mungall, C. J., Torniai, C., Gkoutos, G. V., Lewis, S. E. & Haendel, M. A. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 13, R5 (2012).
Schriml, L. M. & Mitraka, E. The Disease Ontology: fostering interoperability between biological and clinical human disease-related data. Mamm. Genome 26, 584–589 (2015).
Scheubert, K. et al. Significance estimation for large scale metabolomics annotations by spectral matching. Nat. Commun. 8, 1494 (2017).
Sumner, L. W. et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007).
Vázquez-Baeza, Y., Pirrung, M., Gonzalez, A. & Knight, R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2, 1–4 (2013).
Hammons, J. L., Jordan, W. E., Stewart, R. L., Taulbee, J. D. & Berg, R. W. Age and diet effects on fecal bile acids in infants. J. Pediatr. Gastroenterol. Nutr. 7, 30–38 (1988).
Robertson, R. C., Manges, A. R., Finlay, B. B. & Prendergast, A. J. The human microbiome and child growth—first 1000 days and beyond. Trends Microbiol. 27, 131–147 (2019).
Martin, C. B., Hales, C. M., Gu, Q. & Ogden, C. L. Prescription drug use in the United States, 2015–2016 (NCHS Data Brief no. 334) https://www.cdc.gov/nchs/products/databriefs/db334.htm (2019).
Omura, S. et al. A new alkaloid Am-2282 of Streptomyces origin. Taxonomy, fermentation, isolation and preliminary characterization. J. Antibiot. 30, 275–282 (1977).
Peypoux, F., Bonmatin, J. M. & Wallach, J. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 51, 553–563 (1999).
Ernst, M. et al. Molnetenhancer: enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 9, 144 (2019).
Wynalda, M. A., Hutzler, J. M., Koets, M. D., Podoll, T. & Wienkers, L. C. In vitro metabolism of clindamycin in human liver and intestinal microsomes. Drug Metab. Dispos. 31, 878–887 (2003).
Protsyuk, I. et al. 3D molecular cartography using LC–MS facilitated by Optimus and ’ili software. Nat. Protoc. 13, 134–154 (2018).
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
We thank the individuals involved in the funding, administration, sample collection and data acquisition of the public data used in ReDU. We recognize the financial support of the US National Institutes of Health (P41 GM103484, R03 CA211211, R01 LM013115 and R01 GM107550), the Sloan Foundation (R.K.), the Gordon and Betty Moore Foundation (P.C.D., N.B., K.L.M.), a FAPESP fellowship (2018/24865-4), the American Society for Mass Spectrometry (A.K.J.), NSF grants IOS-1656481 (P.C.D. and A.M.C.R.) and ABI-1759980, Netherlands eScience Center no. ASDI.2017.030 (J.J.J.v.d.H.), the Krupp Endowed Fund (R. Coras), the US Office of Naval Research (N00014-15-1-2809) and the University of California, San Diego, Center for Microbiome Innovation SEED grants.
Author information
Authors and Affiliations
Contributions
A.K.J., M.W. and P.C.D. developed the ReDU concept. A.K.J., M.W. and C.M.A. wrote code and engineered the ReDU infrastructure. A.K.J., C.M.A., R.S.A., S.A., A.A.A., G.A., A.T.A., A. Bauermeister, S.B., A. Bouslimani, A.M.C.R., R. Chaar, R. Coras, E.O.E., J.J.J.v.d.H., J.M.G., E.C.G., M.H., K.L.J., Z.K., A.L.G., A.L., L.-I.M., K.L.M., M.J.M., A.V.M., R.C.M., Y.A.M.G., N.H.N., L.F.N., M.E., M.N.E., M.P., D.P., R.Q., N.S., F.V., A.V. and K.C.W. curated metadata enabling ReDU. A.K.J., M.W., C.M.A., S.A.J., L.-I.M., M.E., J.J.J.v.d.H., J.M.G., M.P. and P.C.D. tested the ReDU infrastructure and provided feedback. A.K.J., M.W., C.M.A., M.E., J.J.J.v.d.H., R.K., N.B. and P.C.D. wrote and edited the manuscript. R.K., N.B. and P.C.D. provided supervision and funding support.
Corresponding author
Ethics declarations
Competing interests
P.C.D. is a scientific advisor for Sirenas LLC, Galileio and Cybele Microbiome and scientific advisor and co-founder of Enveda. M.W. is a founder of Ometa Labs LLC. A.A. is a consultant for Ometa Labs LLC.
Additional information
Peer review information Allison Doerr was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Repository-scale molecular cartography enabled by ReDU.
ReDU samples with latitude and longitude information, n = 34,003, were grouped by latitude and longitude (n = 2068 different locations) and plotted colored by number of annotations per file (log10 scaled).
Extended Data Fig. 2 Comparison of bacterial cultures using Group Comparator in ReDU.
Bacterial cultures of 1280|Staphylococcus aureus (n = 49), 1423|Bacillus subtilis (n = 89), and 1883|Streptomyces (n = 7) were compared and chemical differences are illustrated by pyroGlu-Ile, staurosporine, and surfactin-C14.
Extended Data Fig 3 Repository-scale molecular networking of human blood (n = 711), fecal (n = 5,097), and urine (n = 307) supplemented by MolNetEnhancer.
(a) MolNetEnhancer enhanced molecular network in which components are colored based on Classyfire chemical class prediction. (b) Number of nodes per Classyfire chemical class prediction. Nodes without a match in Classyfire are not displayed.
Extended Data Fig 4 Molecular cartography of the distribution of drugs on the human body visualized using ili.
Descaladinose azithromycin, a drug metabolite of azithromycin, distribution in human (n = 17,117; normalized by the number of files per sample).
Supplementary information
Supplementary Information
Supplementary Discussion, Supplementary Table 1 and Supplementary Figs. 1–5.
Supplementary Table 2
Tabulated information for proposed clindamycin metabolites based on repository-scale molecular networking in ReDU.
Supplementary Video 1
The empirically observed UBERON-based location of drugs and drug metabolites observed in metabolomics data 28 August 2019–10 July 2020 processed through ReDU.
Rights and permissions
About this article
Cite this article
Jarmusch, A.K., Wang, M., Aceves, C.M. et al. ReDU: a framework to find and reanalyze public mass spectrometry data. Nat Methods 17, 901–904 (2020). https://doi.org/10.1038/s41592-020-0916-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-020-0916-7
This article is cited by
-
Chemical characteristics vectors map the chemical space of natural biomes from untargeted mass spectrometry data
Journal of Cheminformatics (2025)
-
A guide to reverse metabolomics—a framework for big data discovery strategy
Nature Protocols (2025)
-
Two-layer homolog network approach for PFAS nontarget screening and retrospective data mining
Nature Communications (2025)
-
Introducing Korea metabolomics data repository (KMAP): bridging Korean metabolomics data to global data sharing infrastructure
Metabolomics (2025)
-
Enabling pan-repository reanalysis for big data science of public metabolomics data
Nature Communications (2025)