Abstract
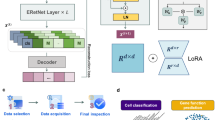
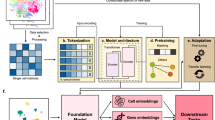
Large pretrained models have become foundation models leading to breakthroughs in natural language processing and related fields. Developing foundation models for deciphering the ‘languages’ of cells and facilitating biomedical research is promising yet challenging. Here we developed a large pretrained model scFoundation, also named ‘xTrimoscFoundationα’, with 100 million parameters covering about 20,000 genes, pretrained on over 50 million human single-cell transcriptomic profiles. scFoundation is a large-scale model in terms of the size of trainable parameters, dimensionality of genes and volume of training data. Its asymmetric transformer-like architecture and pretraining task design empower effectively capturing complex context relations among genes in a variety of cell types and states. Experiments showed its merit as a foundation model that achieved state-of-the-art performances in a diverse array of single-cell analysis tasks such as gene expression enhancement, tissue drug response prediction, single-cell drug response classification, single-cell perturbation prediction, cell type annotation and gene module inference.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data used in this study are publicly available and the usages are illustrated in the Methods. The pretraining datasets were mainly downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/), Single Cell Portal (https://singlecell.broadinstitute.org/single_cell), HCA (https://data.humancellatlas.org/) and EMBL-EBI (https://www.ebi.ac.uk/), and the detailed dataset list we used is in Supplementary Data 1 and 2. The datasets used for downstream tasks can be downloaded from the following links: Baron dataset (https://github.com/mohuangx/SAVER-paper); Zheng68K dataset (https://www.dropbox.com/sh/w3yg2nucnng5v1u/AAAM8Ym_KU9XF4z51RT81eNEa?dl=0); Segerstolpe dataset (https://zenodo.org/records/3357167); CDR dataset (https://github.com/kimmo1019/DeepCDR); Single cell drug response classification dataset (https://github.com/CompBioT/SCAD); Perturbation dataset (https://github.com/snap-stanford/GEARS); Simulated reference and query dataset used for cell mapping (https://doi.org/10.6084/m9.figshare.21456645.v4); and Organoid and in vivo data used for cell mapping (https://doi.org/10.17632/sm67hr5bpm.1). The processed gene expression data and the embeddings generated by scFoundation can be found in our GitHub repository (https://github.com/biomap-research/scFoundation) and figshare (https://doi.org/10.6084/m9.figshare.24049200) (ref. 74).
Code availability
The code for using the online API, the model codes and weight, a demonstration of inferring embeddings, codes of producing the results for the downstream tasks are at the GitHub repository at https://github.com/biomap-research/scFoundation or Zenodo75. A summary of all code and data information is in Supplementary Data 3.
References
Srivastava, A. et al. Beyond the imitation game: quantifying and extrapolating the capabilities of language models. Preprint at arXiv https://doi.org/10.48550/arXiv.2206.04615 (2023).
Jovic, D. et al. Single-cell RNA sequencing technologies and applications: a brief overview. Clin. Transl. Med. 12, e694 (2022).
Regev, A. et al. The Human Cell Atlas. eLife 6, e27041 (2017).
Chen, S. et al. hECA: the cell-centric assembly of a cell atlas. iScience 25, 104318 (2022).
Snyder, M. P. et al. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
The Tabula Sapiens Consortium. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022).
Li, M. et al. DISCO: a database of deeply integrated human single-cell omics data. Nucleic Acids Res. 50, D596–D602 (2022).
Papatheodorou, I. et al. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 48, D77–D83 (2020).
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018).
Brown, T. B. et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 33, 1877–1901 (2020).
Zhao, W. X. et al. A survey of large language models. Preprint at arXiv https://doi.org/10.48550/arXiv.2303.18223 (2023).
Zhang, R., Luo, Y., Ma, J., Zhang, M. & Wang, S. scPretrain: multi-task self-supervised learning for cell-type classification. Bioinformatics 38, 1607–1614 (2022).
Yang, F. et al. scBERT as a large-scale pretrained deep language model for cell type annotation of single-cell RNA-seq data. Nat. Mach. Intell. 4, 852–866 (2022).
Cui, H., Wang, C., Maan, H. & Wang, B. scGPT: towards building a foundation model for single-cell multi-omics using generative AI. Nat Methods https://doi.org/10.1038/s41592-024-02201-0 (2024).
Theodoris, C. V. et al. Transfer learning enables predictions in network biology. Nature https://doi.org/10.1038/s41586-023-06139-9 (2023).
Choromanski, K. et al. Rethinking attention with performers. Preprint at arXiv https://doi.org/10.48550/arXiv.2009.14794 (2022).
Ma, X. et al. Luna: Linear Unified Nested Attention. Adv. Neural Inf. Process. Syst. 34, 2441–2453 (2021).
Gong, J. et al. xTrimoGene: an efficient and scalable representation learner for single-cell RNA-seq data. Preprint at bioRxiv https://doi.org/10.1101/2023.03.24.534055 (2023).
Chen, J. et al. Transformer for one stop interpretable cell type annotation. Nat. Commun. 14, 223 (2023).
He, K. et al. in Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 16000–16009 (IEEE, 2022).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. in Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics 4171–4186 (ACL, 2019).
Edgar, R., Domrachev, M. & Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Seal, R. L. et al. Genenames.org: the HGNC resources in 2023. Nucleic Acids Res. 51, D1003–D1009 (2023).
Kaplan, J. et al. Scaling laws for neural language models. Preprint at arXiv https://doi.org/10.48550/arXiv.2001.08361 (2020).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27 (2018).
Huang, M. et al. SAVER: gene expression recovery for single-cell RNA sequencing. Nat. Methods 15, 539–542 (2018).
Li, W. V. & Li, J. J. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat. Commun. 9, 997 (2018).
Kedzierska, K. Z., Crawford, L., Amini, A. P. & Lu, A. X. Assessing the limits of zero-shot foundation models in single-cell biology. Preprint at bioRxiv https://doi.org/10.1101/2023.10.16.561085 (2023).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Abdelaal, T. et al. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 20, 194 (2019).
Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022).
Polański, K. et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965 (2020).
Unger, F. T., Witte, I. & David, K. A. Prediction of individual response to anticancer therapy: historical and future perspectives. Cell. Mol. Life Sci. 72, 729–757 (2015).
Liu, Q., Hu, Z., Jiang, R. & Zhou, M. DeepCDR: a hybrid graph convolutional network for predicting cancer drug response. Bioinformatics 36, i911–i918 (2020).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Iorio, F. et al. A landscape of pharmacogenomic interactions in cancer. Cell 166, 740–754 (2016).
Bellamy, D., Celi, L. & Beam, A. L. Evaluating progress on machine learning for longitudinal electronic healthcare data. Preprint at arXiv https://doi.org/10.48550/arXiv.2010.01149 (2020).
Geeleher, P., Cox, N. J. & Huang, R. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 15, R47 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Saddoughi, S. A., Song, P. & Ogretmen, B. in Lipids in Health and Disease (eds Quinn, P. J. & Wang, X.) 413–440 (Springer, 2008).
Kurundkar, D. et al. Vorinostat, an HDAC inhibitor attenuates epidermoid squamous cell carcinoma growth by dampening mTOR signaling pathway in a human xenograft murine model. Toxicol. Appl. Pharmacol. 266, 233–244 (2013).
Park, H. et al. Phase I dose-escalation study of the mTOR inhibitor sirolimus and the HDAC inhibitor vorinostat in patients with advanced malignancy. Oncotarget 7, 67521–67531 (2016).
Zibelman, M. et al. Phase I study of the mTOR inhibitor ridaforolimus and the HDAC inhibitor vorinostat in advanced renal cell carcinoma and other solid tumors. Invest. N. Drugs 33, 1040–1047 (2015).
Vasudevan, S. et al. Drug-induced resistance and phenotypic switch in triple-negative breast cancer can be controlled via resolution and targeting of individualized signaling signatures. Cancers 13, 5009 (2021).
Zheng, Z. et al. Enabling single-cell drug response annotations from bulk RNA-seq using SCAD. Adv. Sci. 10, e2204113 (2023).
Ho, Y.-J. et al. Single-cell RNA-seq analysis identifies markers of resistance to targeted BRAF inhibitors in melanoma cell populations. Genome Res. 28, 1353–1363 (2018).
Kinker, G. S. et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 52, 1208–1218 (2020).
Rood, J. E., Maartens, A., Hupalowska, A., Teichmann, S. A. & Regev, A. Impact of the Human Cell Atlas on medicine. Nat. Med. 28, 2486–2496 (2022).
Adamson, B. et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882 (2016).
Dixit, A. et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866 (2016).
Roohani, Y., Huang, K. & Leskovec, J. Predicting transcriptional outcomes of novel multigene perturbations with GEARS. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01905-6 (2023).
Lotfollahi, M., Wolf, F. A. & Theis, F. J. scGen predicts single-cell perturbation responses. Nat. Methods 16, 715–721 (2019).
Lotfollahi, M. et al. Learning interpretable cellular responses to complex perturbations in high-throughput screens. Preprint at bioRxiv https://doi.org/10.1101/2021.04.14.439903 (2021).
Lotfollahi, M. et al. Predicting cellular responses to complex perturbations in high-throughput screens. Mol. Syst. Biol. 19, e11517 (2023).
Segerstolpe, Å. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607 (2016).
Domínguez Conde, C. et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 376, eabl5197 (2022).
Xu, C. et al. Probabilistic harmonization and annotation of single-cell transcriptomics data with deep generative models. Mol. Syst. Biol. 17, e9620 (2021).
Ma, F. & Pellegrini, M. ACTINN: automated identification of cell types in single cell RNA sequencing. Bioinformatics 36, 533–538 (2020).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Tan, Y. & Cahan, P. SingleCellNet: a computational tool to classify single cell RNA-seq data across platforms and across species. Cell Syst. 9, 207–213 (2019).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Date, D. et al. Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J. Biol. Chem. 289, 10318–10329 (2014).
Willis, S. N. et al. Environmental sensing by mature B cells is controlled by the transcription factors PU.1 and SpiB. Nat. Commun. 8, 1426 (2017).
Vasilevsky, N. A., Ruby, C. E., Hurlin, P. J. & Weinberg, A. D. OX40 engagement stabilizes Mxd4 and Mnt protein levels in antigen-stimulated T cells leading to an increase in cell survival. Eur. J. Immunol. 41, 1024–1034 (2011).
Ma, S. et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183, 1103–1116 (2020).
Chen, S., Lake, B. B. & Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 37, 1452–1457 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Beltagy, I., Peters, M. E. & Cohan, A. Longformer: the long-document transformer. Preprint at arXiv https://doi.org/10.48550/arXiv.2004.05150 (2020).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Norman, T. M. et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793 (2019).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128 (2013).
Hao, M. scFoundation: large scale foundation model on single-cell transcriptomics - processed datasets. figshare. https://doi.org/10.6084/m9.figshare.24049200.v3 (2023).
Hao, M. code of scFoundation: large scale foundation model on single-cell transcriptomics. Zenodo https://doi.org/10.5281/zenodo.8330924 (2023).
Acknowledgements
We thank Q. Yin, L. Chao and Z. He from Biomap and Y. Chen, C. Li, H. Bian, J. Li, T. Ma, L. Wei and R. Jiang from Bioinfo Division, Tsinghua University for discussions and comments. This work was partially supported by the National Key R&D Program of China (grant 2021YFF1200901), National Natural Science Foundation of China (NSFC) (grants 62250005 and 61721003) and Tsinghua-Fuzhou Institute for Data Technology (TFIDT2021005).
Author information
Authors and Affiliations
Contributions
M.H., J.M., L.S. and X. Zhang conceived the study. M.H. X. Zeng and Y.G. collected the downstream datasets involved in this article. Y.G. and L.S. developed data collection criteria and strategies for pretraining. M.H., J.G., X. Zeng, C.L., T.W. and X.C. proposed the pretraining framework. M.H., J.G., X. Zeng and C.L. implemented and pretrained the models. M.H. and J.G. benchmarked all methods. J.G., X. Zeng, C.L., T.W., X.C., J.M., L.S. and X. Zhang provided advice on pretraining framework design and downstream tasks. M.H., J.G., J.M., L.S. and X. Zhang wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.G., X.Ze., C.L, Y.G., X.C., T.W. and L.S. are employees of BioMap. M.H. contributed to this work while part-time interning at BioMap. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Lin Tang, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–13, Figs. 1–16 and Tables 1–10.
Supplementary Data 1
Name of datasets used for pretraining.
Supplementary Data 2
Corresponding study names of datasets used for pretraining.
Supplementary Data 3
A summary of all code and data information.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hao, M., Gong, J., Zeng, X. et al. Large-scale foundation model on single-cell transcriptomics. Nat Methods 21, 1481–1491 (2024). https://doi.org/10.1038/s41592-024-02305-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-024-02305-7
This article is cited by
-
Cellular neighborhoods in cancer
Nature Cancer (2026)
-
Discovery of predictive biomarkers for cancer therapy through computational approaches
Nature Reviews Clinical Oncology (2026)
-
Interpretation, extrapolation and perturbation of single cells
Nature Reviews Genetics (2026)
-
A unified time-frequency foundation model for sleep decoding
Nature Communications (2026)
-
Sex differences in bladder cancer: understanding biological and clinical implications
Biology of Sex Differences (2025)