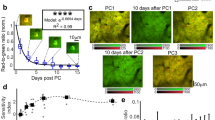
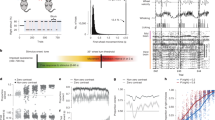
Abstract
Technologies that can record neural activity at cellular resolution at multiple spatial and temporal scales are typically much larger than the animals that are being recorded from and are thus limited to recording from head-fixed subjects. Here we have engineered robotic neural recording devices—‘cranial exoskeletons’—that assist mice in maneuvering recording headstages that are orders of magnitude larger and heavier than the mice, while they navigate physical behavioral environments. We discovered optimal controller parameters that enable mice to locomote at physiologically realistic velocities while maintaining natural walking gaits. We show that mice learn to work with the robot to make turns and perform decision-making tasks. Robotic imaging and electrophysiology headstages were used to record recordings of Ca2+ activity of thousands of neurons distributed across the dorsal cortex and spiking activity of hundreds of neurons across multiple brain regions and multiple days, respectively.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets reported here are openly available via the Zenodo repository at https://doi.org/10.5281/zenodo.10454727 (ref. 49). Source data are provided with this paper.
Code availability
All code for controlling the robot and for data analysis is available via the GitHub repository at https://github.com/bsbrl/exoskeleton. Software is made available via CC BY-NC-ND license.
References
Jun, J. J. et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236 (2017).
Steinmetz, N. A., Zatka-Haas, P., Carandini, M. & Harris, K. D. Distributed coding of choice, action and engagement across the mouse brain. Nature 576, 266–273 (2019).
Steinmetz, N. A. et al. Neuropixels 2.0: a miniaturized high-density probe for stable, long-term brain recordings. Science 372, eabf4588 (2021).
Siegle, J. H. et al. Survey of spiking in the mouse visual system reveals functional hierarchy. Nature 592, 86–92 (2021).
Stirman, J. N., Smith, I. T., Kudenov, M. W. & Smith, S. L. Wide field-of-view, multi-region, two-photon imaging of neuronal activity in the mammalian brain. Nat. Biotechnol. 34, 857–862 (2016).
Sofroniew, N. J., Flickinger, D., King, J. & Svoboda, K. A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. eLife https://doi.org/10.7554/eLife.14472 (2016).
Yu, C. H., Stirman, J. N., Yu, Y., Hira, R. & Smith, S. L. Diesel2p mesoscope with dual independent scan engines for flexible capture of dynamics in distributed neural circuitry. Nat. Commun. 12, 1–8 (2021).
Ghanbari, L. et al. Cortex-wide neural interfacing via transparent polymer skulls. Nat. Commun. 10, 1–13 (2019).
Rynes, M. L. et al. Miniaturized head-mounted microscope for whole-cortex mesoscale imaging in freely behaving mice. Nat. Methods 18, 417–425 (2021).
Harvey, C. D., Collman, F., Dombeck, D. A. & Tank, D. W. Intracellular dynamics of hippocampal place cells during virtual navigation. Nature 461, 941–946 (2009).
Saleem, A. B., Ayaz, A. I., Jeffery, K. J., Harris, K. D. & Carandini, M. Integration of visual motion and locomotion in mouse visual cortex. Nat. Neurosci. 16, 1864–1869 (2013).
Murphy, T. H. et al. High-throughput automated home-cage mesoscopic functional imaging of mouse cortex. Nat. Commun. 7, 1–12 (2016).
Scott, B. B., Brody, C. D. & Tank, D. W. Cellular resolution functional imaging in behaving rats using voluntary head restraint. Neuron 80, 371–384 (2013).
Go, M. A. et al. Place cells in head-fixed mice navigating a floating real-world environment. Front. Cell Neurosci. 15, 19 (2021).
Voigts, J. & Harnett, M. T. Somatic and dendritic encoding of spatial variables in retrosplenial cortex differs during 2D navigation. Neuron 105, 237–245.e4 (2020).
Minderer, M., Harvey, C. D., Donato, F. & Moser, E. I. Virtual reality explored. Nature 533, 324–325 (2016).
Meyer, A. F., O’Keefe, J. & Poort, J. Two distinct types of eye-head coupling in freely moving mice. Curr. Biol. 30, 2116–2130.e6 (2020).
Aghajan, Z. M. et al. Impaired spatial selectivity and intact phase precession in two-dimensional virtual reality. Nat. Neurosci. 18, 121–128 (2014).
Ghosh, K. K. et al. Miniaturized integration of a fluorescence microscope. Nat. Methods 8, 871–878 (2011).
Cai, D. J. et al. A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534, 115–118 (2016).
Voigts, J., Siegle, J., Pritchett, D. L. & Moore, C. I. The flexDrive: an ultra-light implant for optical control and highly parallel chronic recording of neuronal ensembles in freely moving mice. Front. Syst. Neurosci. 7, 8 (2013).
Meyer, A. F., Poort, J., O’Keefe, J., Sahani, M. & Linden, J. F. A head-mounted camera system integrates detailed behavioral monitoring with multichannel electrophysiology in freely moving mice. Neuron 100, 46–60.e7 (2018).
Keemink, A. Q. L., van der Kooij, H. & Stienen, A. H. A. Admittance control for physical human–robot interaction. Int. J. Rob. Res. 37, 1421–1444 (2018).
Mathis, A. et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018).
Broom, L. et al. A translational approach to capture gait signatures of neurological disorders in mice and humans. Sci. Rep. 7, 1–17 (2017).
Donaldson, P. D. et al. Polymer skulls with integrated transparent electrode arrays for cortex-wide opto-electrophysiological recordings. Adv. Health. Mater. 11, 2200626 (2022).
Franco, S. J. et al. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science 337, 746–749 (2012).
Daigle, T. L. et al. A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174, 465–480.e22 (2018).
Kauvar, I. V. et al. Cortical observation by synchronous multifocal optical sampling reveals widespread population encoding of actions. Neuron 107, 351–367.e19 (2020).
Pachitariu, M. et al. Suite2p: beyond 10,000 neurons with standard two-photon microscopy. Preprint at bioRxiv https://doi.org/10.1101/061507 (2017).
Stringer, C. et al. Spontaneous behaviors drive multidimensional, brainwide activity. Science 364, eaav7893 (2019).
Musall, S., Kaufman, M. T., Juavinett, A. L., Gluf, S. & Churchland, A. K. Single-trial neural dynamics are dominated by richly varied movements. Nat. Neurosci. 22, 1677–1686 (2019).
West, S. L. et al. Wide-field calcium imaging of dynamic cortical networks during locomotion. Cereb. Cortex 32, 2668–2687 (2022).
Pachitariu, M., Sridhar, S., Pennington, J. & Stringer, C. Spike sorting with Kilosort4. Nat. Methods 21, 914–921 (2024).
Rossant, C. et al. Spike sorting for large, dense electrode arrays. Nat. Neurosci. 19, 634–641 (2016).
Ebrahimi, S. et al. Emergent reliability in sensory cortical coding and inter-area communication. Nature 605, 713–721 (2022).
Wiltschko, A. B. et al. Mapping sub-second structure in mouse behavior. Neuron 88, 1121–1135 (2015).
Markowitz, J. E. et al. The striatum organizes 3D behavior via moment-to-moment action selection. Cell 174, 44–58.e17 (2018).
Sekiguchi, S. et al. Uncertainty-aware non-linear model predictive control for human-following companion robot. In Proc. IEEE International Conference on Robotics and Automation https://doi.org/10.1109/ICRA48506.2021.9561974, 8316–8322 (IEEE, 2021).
Wang, L., Van Asseldonk, E. H. F. & Van Der Kooij, H. Model predictive control-based gait pattern generation for wearable exoskeletons. In IEEE International Conference on Rehabilitation Robotics https://doi.org/10.1109/ICORR.2011.5975442 (IEEE, 2011).
Demas, J. et al. High-speed, cortex-wide volumetric recording of neuroactivity at cellular resolution using light beads microscopy. Nat. Methods 18, 1103–1111 (2021).
Peters, A. J., Fabre, J. M. J., Steinmetz, N. A., Harris, K. D. & Carandini, M. Striatal activity topographically reflects cortical activity. Nature 591, 420–425 (2021).
Carrillo-Reid, L., Han, S., Yang, W., Akrouh, A. & Yuste, R. Controlling visually guided behavior by holographic recalling of cortical ensembles. Cell 178, 447–457.e5 (2019).
Szalay, G. et al. Fast 3D imaging of spine, dendritic, and neuronal assemblies in behaving animals. Neuron 92, 723–738 (2016).
Rynes, M. L. et al. Assembly and operation of an open-source, computer numerical controlled (CNC) robot for performing cranial microsurgical procedures. Nat. Protoc. 15, 1992–2023 (2020).
Hattori, R. & Komiyama, T. Longitudinal two-photon calcium imaging with ultra-large cranial window for head-fixed mice. STAR Protoc. 3, 101343 (2022).
Paxinos, G. & Franklin, K. B. J. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates (Academic Press, 2019).
Hilafu, H., Safo, S. E. & Haine, L. Sparse reduced-rank regression for integrating omics data. BMC Bioinform. 21, 1–17 (2020).
Hope, J. et al. Dataset for 'Brain-wide neural recordings in mice navigating physical spaces enabled by a cranial exoskeleton'. Zenodo https://doi.org/10.5281/zenodo.10454727 (2023).
Acknowledgements
We acknowledge the staff at Research Animal Resources, University of Minnesota, for animal care and housing; J. O’Brien for his initial exploration of the exoskeleton concept; D. Surinach for assistance training models in DeepLabCut; J. Hu for assistance using Suite2p and processing ΔF/F traces; E. Troester for assistance with code for video acquisition; A. Cherkkil for consultation on mesoscale imaging headstage components. Funding sources: University of Minnesota Mechanical Engineering Department, Minnesota's Discovery, Research, and InnoVation Economy (MnDRIVE) Robotics, sensors, and advanced manufacturing in Minnesota (RSAM), the McKnight Foundation, the Minnesota Robotics Institute, National Institutes of Health grant R01NS11128, and BRAIN Initiative grants RF1NS113287 to S.K., P30DA048742 and RF1NS126044 grants to S.K. and T.E.
Author information
Authors and Affiliations
Contributions
J.H. and S.K. conceptualized the exoskeleton. J.H., P.-H.C. and Z.V. designed and built the exoskeleton hardware. J.H. developed the exoskeleton software. J.H., P.-H.C., S.F. and I.H. performed animal surgeries. J.H., T.B., Z.V. and K.S. conducted animal behavior training. J.H., P.-H.C. and K.S. performed exoskeleton development experiments. J.H. and E.K. performed imaging experiments. J.H. and T.B. performed neural probe experiments. J.H., T.B., M.F. and R.C. processed raw experimental data. J.H., T.B., M.F. and S.K. performed data analysis. J.H., T.B., M.F. and S.K. visualized data. J.H., T.B., P.-H.C. and S.K. wrote the original manuscript draft. J.H., T.B., R.C., T.E. and S.K. reviewed and edited the manuscript. J.H. and S.K. supervised all aspects of this work. T.E. and S.K. acquired funding.
Corresponding author
Ethics declarations
Competing interests
S.K. is co-founder of Objective Biotechnology Inc. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Diego Restrepo, Simon Schultz, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling editor Nina Vogt, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Velocity – acceleration profiles of freely behaving mice.
(a) Schematic of the method used to calculate velocities and accelerations in the mouse’s coordinate frame from marker-less tracking data. (b) Example data of velocity and acceleration time series in the mouse’s x axis (forwards/backwards). (c) Velocity-acceleration profile of the data in (b). (d) Close-up view of a 20 s clip (blue trace) of the data in (b), with two periods of motion highlighted (orange and yellow traces). (e) Velocity-acceleration profiles of the data in (d). (f) Example data of velocity and acceleration time series in the mouse’s y (lateral) and yaw (rotation left and right) axes (top, y; bottom, yaw). (g) Velocity-acceleration profiles of the data in (f), (top, y; bottom, yaw). (h) Close-up view of a 15 s clip of the data in (f), with two periods of motion highlighted (purple and green traces). (i) Velocity-acceleration profile of the data in (h).
Extended Data Fig. 2 Pitch and height configuration for the exoskeleton.
(a) Schematic of the methodology used to configure the pitch and height settings of the exoskeleton. (b) Images of a freely behaving mouse overlaid with digitally labelled points on the left paws and other body parts, moving at 3 different forwards velocities (1., low velocity; 2., medium velocity; 3., high velocity). (c) Left - Plot of head pitch angles and velocity for all mice (n = 4), with a quadratic fit to the data (red) and the pitch value implemented on the exoskeleton (blue). Numbers correspond to images in (b). Middle - Plot of snout height and velocity for all mice (n = 4), with a quadratic fit to the data (red) and the height value implemented on the exoskeleton (blue). Numbers correspond to images in (b). Right - Plot of the pitch angle and snout height for all mice (n = 4), with a quadratic fit to the data (red) and the height value implemented on the exoskeleton (blue).
Extended Data Fig. 3 Tuning the admittance controller in the mouse’s x axis.
(a) Image of a mouse maneuvering the exoskeleton around the linear oval track used in experiments for tuning the admittance controller in the mouse’s x axis. (b) Plots of raw velocity-acceleration-force data from all mice (n = 4) and the associated admittance plane of the controller, arranged in order of increasing mass (left to right) and damping (bottom to top) for the 14 combinations of values evaluated. (Inset) Legend showing example plot with labelled axes and color-scale of the admittance plane (or manifold). (c) Example data of the mouse’s path around the oval track, where all data within the oval track was used to evaluate admittance tuning. (d) Data bounds of the velocity-acceleration profiles of freely behaving mice (grey; 3 mice; 6 sessions) and of mice maneuvering the exoskeleton with low (top-left), tuned (top-center), and high (top-right) mass and damping values (blue; 4 mice; 4 sessions), and with tuned mass and damping values after training (bottom-center; blue; 1 mouse; 4 sessions). (e) Distribution of the velocity peak amplitudes (top left, n = 2,822 samples) and acceleration peak amplitudes (top right, n = 5,027 samples), and swarm plots of all velocity amplitudes (bottom left, n = 242,490 samples) and acceleration amplitudes (bottom right, n = 242,490 samples) of individual mice, when freely behaving, and when maneuvering the exoskeleton with low, tuned, and high mass and damping values, and tuned mass and damping values after training, boxplots indicate minimum and maximum outliers as circles, minimum and maximum inliers as whiskers, 25th and 75th percentiles as solid boxes, and median values as a black point in a white circle. (f) Mean and 95% confidence intervals of all velocity (left) and acceleration (right) peaks, with p-values calculated using 1-way ANOVA with n = 100 representative sample peaks per group, n.s. p > 0.05, * p < 0.05, ** p < 0.01. All-to-all velocity p-values: FB-Low p ≈ 0, FB-Tuned p ≈ 0, FB-high p ≈ 0, FB-T&T p = 0.03, Low-Tuned p = 1.9×10-4, Low-High p ≈ 0, Low-T&T p ≈ 0, Tuned-High p ≈ 0, Tuned-T&T p ≈ 0, High-T&T p ≈ 0. All-to-all acceleration p-values: FB-Low p = 6.9×10-13, FB-Tuned p ≈ 0, FB-high p ≈ 0, FB-T&T p = 0.04, Low-Tuned p ≈ 0, Low-High p = 6.5×10-17, Low-T&T p = 3.6×10-5, Tuned-High p ≈ 0, Tuned-T&T p ≈ 0, High-T&T p ≈ 0.
Extended Data Fig. 4 Tuning the admittance controller in the mouse’s y and yaw axes.
(a) Image of a mouse maneuvering the exoskeleton locomoting around the 8-maze arena used in experiments for tuning the admittance controller in the mouse’s y and yaw axes. (b) Example data of the mouse’s path around the 8-maze arena, showing the turning zone in the center of the arena which was used to evaluate admittance tuning. (c) Plots of raw data velocity-acceleration-force data in the mouse’s y axis for all mice (n = 2) and the associated admittance plane of the controller, arranged in order of increasing mass (left to right) and damping (bottom to top) for the 7 combinations of values evaluated. Legend (inset) showing example plot with labelled axes and color-scale of the admittance plane (or manifold). (d) As in (c) but for the 6 combinations of mass and damping evaluated in the mouse’s yaw axis. (e) Data bounds of velocity-acceleration profiles in the mouse’s y-axis for freely behaving mice (grey; 3 mice; 6 sessions, n = 4,711 samples) and for mice maneuvering the exoskeleton with low (left, n = 3,139 samples), medium (center, n = 6,921 samples), and high (right, n = 8,835 samples) mass and damping values (blue; 2 mice; 2 sessions). (f) As in (e) but for the mouse’s yaw axis. (g) Distribution of the velocity peak amplitudes (left, n = 631 samples) and of acceleration peak amplitudes (right, n = 1,917 samples) of individual mice, when freely behaving, and when maneuvering the exoskeleton with low, tuned, and high mass and damping values. Significance (p-values) calculated using 1-way ANOVA between groups, n.s. p > 0.05, * p < 0.05, ** p < 0.01. All-to-all velocity p-values: FB-Low p = 0.29, FB-Mid p = 0.03, FB-High p = 0.03, Low-Mid p = 0.74, Low-High p = 0.72, Mid-High p = 1.0. All-to-all acceleration p-values: FB-Low p = 0.89, FB-Mid p = 0.11, FB-High p = 0.66, Low-Mid p = 0.015, Low-High p = 0.97, Mid-High p = 3.6×10-3. (h) As in (g) but for the mouse’s yaw axis (n = 718 velocity samples, n = 1706 acceleration samples). All-to-all velocity p-values: FB-Low p = 8.8×10-4, FB-Mid p = 0.99, FB-High p = 0.99, Low-Mid p = 2.1×10-4, Low-High p = 2.2×10-4, Mid-High p = 1.0. All-to-all acceleration p-values: FB-Low p = 1.0, FB-Mid p = 0.89, FB-High p = 0.45, Low-Mid p = 0.85, Low-High p = 0.39, Mid-High p = 0.87. All boxplots indicate minimum and maximum values as whiskers, 25th and 75th percentiles as solid boxes, and median values as a black point in a white circle.
Extended Data Fig. 5 Forces during exoskeleton turn-training in the 8-maze arena.
(a) Color coded symbols of the 8-maze arena with both doors in place (turn-training) and mouse maneuvering the exoskeleton indicating the data in the figure. (b) Distribution of force peaks during left (blue) and right (orange) turns through the turning zone of the 8maze for all mice (n = 5) across 8 sessions (top, y forces; bottom, yaw torques), boxplots indicate minimum and maximum values as whiskers, 25th and 75th percentiles as solid boxes, and median values as a black point in a white circle. Shaded regions connect mean values for each session. (c) Change (arrows) in force peaks for each mouse (left) and for all mice (right, n = 5) between their first and final sessions for left (blue) and right (orange) turns (top, y forces, n = 918 samples; bottom, yaw torques, n = 967 samples), plots indicate mean value as a circle with lines showing standard error.
Extended Data Fig. 6 Performance during the decision-making task on the exoskeleton.
(a) Color coded symbols of the 8-maze arena with one door (decision-making) in place and mouse maneuvering the exoskeleton indicating the data in the figure. (b) Paths through the turning zone for correct (green) and incorrect (red) trials for all mice (n = 8) across 8 sessions. (c) The total number of trials per session for individual mice and the mean across all mice (n = 8). (d) Plot of the total number of trials in each session (left-axis; dashed black line) and the number of decision trials in each session (left-axis; solid black line) where decision trials are less than total trials because mice had 2 or more trials at the start of each session with the training door in place in the 8-maze arena. The number of mice in each session (right-axis; blue line). (e) Mean and 95% confidence intervals of the path tortuosity during decision trials for all mice (n = 8; green, correct decisions; red, incorrect decisions). (f) Decision-making performance for each mouse while maneuvering the exoskeleton.
Extended Data Fig. 7 Summary of mesoscale imaging results across all mice.
(a) Symbols of the 8-maze arena with both doors in place (turn-training) and mouse maneuvering the mesoscale imaging headstage indicating the data in the figure. (b) Anatomical locations of all cells imaged in each mouse during an example session in the 8-maze arena. (c) Atlas of the mouse cortex with color coded functional regions. (d) Fluorescence (ΔF/F) distribution for each mouse (n = 2383, 2515, 481, 386, respectively) broken down by brain region, boxplots indicate minimum and maximum outliers as circles, minimum and maximum inliers as whiskers, 25th and 75th percentiles as solid boxes, and median values as a black point in a white circle. (e) Number of cells imaged in each mouse broken down by region. (f) Kernel matrix of normalized Beta weights for each mouse, obtained using linear regression on the inferred spike rate of each cell and the location of the mouse in the 8-maze arena (82 location bins), with rows in the kernel sorted by onset of the maximum Beta weight.
Extended Data Fig. 8 Kernel matrix cross-validation.
(a) Normalized neural activity maps for each mouse obtained for odd- and even-numbered trials, with rows in the plot sorted by location of maximum activity for each cell in the odd-numbered trails. (b) Histogram of Pearson correlations in Beta weights between odd- and even-numbered trials for each cell (n = 2383, 2515, 481, 386, respectively).
Extended Data Fig. 9 Summary of electrophysiology results across all mice.
(a) Symbols of the 8-maze arena with one door in place (decision-making) and mouse maneuvering the electrophysiology headstage indicating the data in the figure, and schematics showing the locations of the 6 recording sites. (b) Mean spike waveform (black) and a subsample of 50 individual spikes for each cell identified in the electrophysiology recordings in each mouse during the navigational decision-making task. (c) Spike amplitudes of each of the mean waveforms shown in (b). (d) Kernel matrices of normalized Beta weights for mouse 1, obtained using linear regression on the z-scored spike rate of each cell and the location of the mouse in the 8-maze arena (196 location bins). Kernel matrices from all 3 days are concatenated vertically with (left) the recording site labelled and color-coded, and (right) global X-Y paths taken by the mouse through the turning zone for each day. (e) As in (d) but for mouse 4.
Extended Data Fig. 10 Neuropixel headstage design and multisite recordings.
(a) Mouse navigating environment with a headstage loaded with 4 Neuropixel probes. (b) Photograph of the headstage with 4 Neuropixel probes inserted through the top protective cap of the implant. (c) Schematics showing the locations of the 4 insertion sites. (d) Example data across 2 successfully inserted probes showing 1 s of raw voltage signal traces from 5 channels (top), corresponding spike cluster mean waveforms with 100 example traces (bottom-left), and cluster cross correlograms (bottom-right). (e) Mouse behavior in the 8-maze arena (X axis position and velocity) and spike rates of individual clusters extracted from 2 inserted probes. Spike rate is limited up to 50 Hz to increase visibility.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11 and Notes 1–4.
Supplementary Video 1
Video (8× speed) showing top-down view of a mouse in open-field arena with several body points labeled using labeled using DeepLabCut marker-less tracking software (left window), and the corresponding velocity-acceleration profile for forward/backward motion in the mouse’s frame of reference (gray; right window). Transition at half-way to a mouse maneuvering the exoskeleton around the linear oval track with tuned mass and damping values and the corresponding velocity–acceleration profile (blue).
Supplementary Video 2
Video (1× speed) showing a side-on-vide of a freely behaving mouse locomoting along a 27-cm-long straight section of the linear oval track (left window), with left paws and other body points labeled using DeepLabCut marker-less tracking software, and the corresponding gait metrics for steps in the video (large gray dots) overlaid on top of the gait metrics for all mice (right window). Transition at half-way to the same mouse maneuvering the exoskeleton with metrics for steps in the video (large blue dots) overlaid on top of the gait metrics for all mice.
Supplementary Video 3
Video (2× speed) showing a trained mouse performing the navigational decision-making task (sensory cue-guided alternating choice) while maneuvering the exoskeleton. Top half of the video shows (left) view from behavioral camera mounted to the headstage and (top right and bottom right) views from two cameras placed around the behavioral arena. The bottom half of the video shows (top) the force time series in x, y and yaw, and (bottom) the velocity and acceleration profiles in x, y and yaw, where thick lines indicate the mouse is in the turning zone.
Supplementary Video 4
Video (8× speed) showing a mouse completing 16 turns in the eight-maze arena while maneuvering the imaging headstage on the exoskeleton (top-left window), with corresponding video of fluorescence in the mouse’s cortex captured using the imaging headstage (bottom-left window). Single cell activity is visible in the four, 1 × 1 mm, ROIs (right windows). Transition half-way to a maximum intensity image of the brain (bottom-left window), and to plot of the ΔF/F traces from all cells in each ROI (right windows).
Supplementary Video 5
Video (8× speed) showing a mouse completing 16 turns in the eight-maze arena while maneuvering the imaging headstage on the exoskeleton (top-left window), with corresponding video of fluorescence in the mouse’s cortex captured using the imaging headstage (bottom-left window). Single cell activity is visible in the four, 1 × 1 mm, ROIs (right windows). Transition half-way to a maximum intensity image of the brain (bottom-left window), and to plot of the ΔF/F traces from all cells in each ROI (right windows).
Source data
Source Data Fig. 1
Figure plot source data.
Source Data Fig. 2
Figure plot source dat.
Source Data Fig. 3
Figure plot source data.
Source Data Fig. 4
Figure plot source data.
Source Data Fig. 5
Figure plot source data.
Source Data Extended Data Fig. 1
Figure plot source data.
Source Data Extended Data Fig. 2
Figure plot source data.
Source Data Extended Data Fig. 3
Figure plot source data.
Source Data Extended Data Fig. 4
Figure plot source data.
Source Data Extended Data Fig. 5
Figure plot source data.
Source Data Extended Data Fig. 6
Figure plot source data.
Source Data Extended Data Fig. 7
Figure plot source data.
Source Data Extended Data Fig. 8
Figure plot source data.
Source Data Extended Data Fig. 9
Figure plot source data.
Source Data Extended Data Fig. 10
Figure plot source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hope, J., Beckerle, T.M., Cheng, PH. et al. Brain-wide neural recordings in mice navigating physical spaces enabled by robotic neural recording headstages. Nat Methods 21, 2171–2181 (2024). https://doi.org/10.1038/s41592-024-02434-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-024-02434-z
This article is cited by
-
Year in review 2024
Nature Methods (2025)