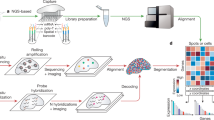
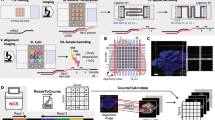
Abstract
Spatiomolecular analyses are key to study tissue functions and malfunctions. However, we lack profiling tools for spatial transcriptomics that are easy to adopt, low cost and scalable in terms of sample size and number. Here, we describe a method, Array-seq, to repurpose classical oligonucleotide microarrays for spatial transcriptomics profiling. We generate Array-seq slides from microarrays carrying custom-design probes that contain common sequences flanking unique barcodes at known coordinates. Then we perform a simple, two-step reaction that produces mRNA capture probes across all spots on the microarray. We demonstrate that Array-seq yields spatial transcriptomes with high detection sensitivity and localization specificity using histological sections from mouse tissues as test systems. Moreover, we show that the large surface area of Array-seq slides yields spatial transcriptomes (i) at high throughput by profiling multi-organ sections, (ii) in three dimensions by processing serial sections from one sample, and (iii) across whole human organs. Thus, by combining classical DNA microarrays and next-generation sequencing, we have created a simple and flexible platform for spatiomolecular studies of small-to-large specimens at scale.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The sequencing data generated during this study have been deposited in the Gene Expression Omnibus under accession numbers GSE266246 (bulk, whole-tissue RNA-seq) and GSE266244 (ST). The uncropped scan of the DNA electrophoresis gel and the H&E images are available via Zenodo at https://doi.org/10.5281/zenodo.13234161. Source data are provided with this paper.
Code availability
All scripts are publicly available at Zenodo via https://doi.org/10.5281/zenodo.10963424.
References
Moses, L. & Pachter, L. Museum of spatial transcriptomics. Nat. Methods 19, 534–546 (2022).
Lein, E., Borm, L. E. & Linnarsson, S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358, 64–69 (2017).
Rao, A., Barkley, D., Franca, G. S. & Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220 (2021).
Crosetto, N., Bienko, M. & van Oudenaarden, A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet. 16, 57–66 (2015).
Stahl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681 (2020).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 (2022).
Cho, C. S. et al. Microscopic examination of spatial transcriptome using Seq-Scope. Cell 184, 3559–3572 (2021).
Fu, X. et al. Polony gels enable amplifiable DNA stamping and spatial transcriptomics of chronic pain. Cell 185, 4621–4633 (2022).
Salmen, F. et al. Barcoded solid-phase RNA capture for Spatial Transcriptomics profiling in mammalian tissue sections. Nat. Protoc. 13, 2501–2534 (2018).
Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 (1995).
Chee, M. et al. Accessing genetic information with high-density DNA arrays. Science 274, 610–614 (1996).
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000).
Islam, S. et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166 (2014).
Hughes, T. R. et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19, 342–347 (2001).
Chen, X., Sun, Y. C., Church, G. M., Lee, J. H. & Zador, A. M. Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res. 46, e22 (2018).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Long, Y. et al. Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST. Nat. Commun. 14, 1155 (2023).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 39, 1375–1384 (2021).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Ma, Y. & Zhou, X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat. Biotechnol. 40, 1349–1359 (2022).
Tepe, B. et al. Single-cell RNA-seq of mouse olfactory bulb reveals cellular heterogeneity and activity-dependent molecular census of adult-born neurons. Cell Rep. 25, 2689–2703 (2018).
Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol. 40, 517–526 (2022).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022).
Kirita, Y., Wu, H., Uchimura, K., Wilson, P. C. & Humphreys, B. D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl Acad. Sci. USA 117, 15874–15883 (2020).
Hildebrandt, F. et al. Spatial transcriptomics to define transcriptional patterns of zonation and structural components in the mouse liver. Nat. Commun. 12, 7046 (2021).
Alexandre, Y. O. & Mueller, S. N. Splenic stromal niches in homeostasis and immunity. Nat. Rev. Immunol. 23, 705–719 (2023).
Cang, Z. et al. Screening cell–cell communication in spatial transcriptomics via collective optimal transport. Nat. Methods 20, 218–228 (2023).
Chen, A. et al. Single-cell spatial transcriptome reveals cell-type organization in the macaque cortex. Cell 186, 3726–3743 (2023).
Schott, M. et al. Open-ST: high-resolution spatial transcriptomics in 3D. Cell 187, 3953–3972 (2024).
Takahama, M. et al. A pairwise cytokine code explains the organism-wide response to sepsis. Nat. Immunol. 25, 226–239 (2024).
Liu, Y. et al. High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. 41, 1405–1409 (2023).
Vickovic, S. et al. SM-Omics is an automated platform for high-throughput spatial multi-omics. Nat. Commun. 13, 795 (2022).
Fan, Y. et al. Expansion spatial transcriptomics. Nat. Methods 20, 1179–1182 (2023).
Buschmann, T. DNABarcodes: an R package for the systematic construction of DNA sample tags. Bioinformatics 33, 920–922 (2017).
Soumillon, M., Cacchiarelli, D., Semrau, S., van Oudenaarden, A. & Mikkelsen, T. Characterization of directed differentiation by high-throughput single-cell RNA-seq. Preprint at bioRxiv https://doi.org/10.1101/003236 (2014).
Pandey, S. et al. A whole-tissue RNA-seq toolkit for organism-wide studies of gene expression with PME-seq. Nat. Protoc. 15, 1459–1483 (2020).
Kadoki, M. et al. Organism-level analysis of vaccination reveals networks of protection across tissues. Cell 171, 398–413 (2017).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
van der Walt, S. et al. scikit-image: image processing in Python. PeerJ 2, e453 (2014).
Ni, Z. et al. SpotClean adjusts for spot swapping in spatial transcriptomics data. Nat. Commun. 13, 2971 (2022).
Fang, Z., Liu, X. & Peltz, G. GSEApy: a comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics https://doi.org/10.1093/bioinformatics/btac757 (2023).
Acknowledgements
We thank members of the laboratory of N.C., V. Maran and M. Soumillon for valuable discussions. N.C. thanks S. Uematsu for the introduction to DNA microarrays. We thank UChicago Core facilities for support: the Single Cell Immunophenotyping Core, the Animal Resources Center, the Research Computing Center and the Integrated Light Microscopy Core; and SciStories for help with artwork. D.C. was supported by National Institutes of Health (NIH) grant T32-GM007281. N.C was supported by NIH grants DP2-AI145100 and U01-AI160418, the CZI grant DAF2020-217464 and a grant from the Chan Zuckerberg Initiative DAF (https://doi.org/10.37921/767230ofotux), an advised fund of Silicon Valley Community Foundation (funder https://doi.org/10.13039/100014989), the Agilent ACT-UR program (grant ID 4843), the UChicago Center for Interdisciplinary Study of Inflammatory Intestinal Disorders (NIDDK P30 DK042086), the UChicago Diabetes Research and Training Center (NIDDK P30 DK020595), the Duckworth Family Commercial Promise Cancer Research Award, the Robert Lavichant Faculty Innovation Award and funds from the Chicago Immunoengineering Innovation Center and the Pritzker School of Molecular Engineering at the University of Chicago.
Author information
Authors and Affiliations
Contributions
N.C. developed the concept. D.C. and N.C. designed the experiments and analysis approaches. D.C. performed the experiments and conducted the analyses. L.M. and T.U. performed optimization experiments. D.C. and N.C. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
D.C. and N.C. are authors on patent PCT/US23/13010 covering the described technology. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Christoph Kuppe, Yang Liu, Neta Milman, Sinem Saka, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Madhura Mukhopadhyay, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Sequence level overview of the on-slide assembly procedure yielding mRNA capture probes.
a, Custom sequence oligonucleotide microarrays containing two common sequences (Anchors 1 and 2) and a spatial barcode sequence unique to each spot on the array are hybridized with indicated oligonucleotides (Step 1). On-slide extension-ligation using a DNA polymerase and a DNA ligase yields a fully assembled mRNA capture probe containing a sequencing adaptor, a spatial barcode, UMIs, and an oligo(dT) sequence (Step 2). In situ reverse transcription in tissue sections placed on an Array-seq slide generates full-length cDNAs with a template switching oligo (TSO) sequence in 3’ (Step 3). Full length cDNAs are eluted from the slides and processed for RNA sequencing library construction (Step 4). b, c, Ligation bias analysis after extension-ligation of mRNA capture probes. Bar plots showing the number of UMIs captured by spot across an Array-seq slide after grouping and normalizing by the last single (b) or two (c) bases of the spatial barcode sequence. Data are presented as mean values ± SD (n = 3490 spots for A, 3756 for C, 2767 for G, and 3771 for T in b; and n = 647 for AA, 1000 for AC, 1000 for AG, 749 for AT, 1057 for CA, 789 for CC, 800 for CG, 1162 for CT, 997 for GA, 823 for GC, 1045 for GT, 789 for TA, 1144 for TC, 967 for TG, 770 for TT in c).
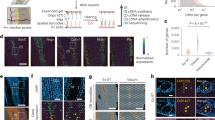
Extended Data Fig. 2 Reproducibility of Array-seq data from mouse main olfactory bulb sections.
a, Correlation (Pearson’s coefficient) between Array-seq replicates 1 and 2 from two sections obtained from two independent mouse main olfactory bulb (MOB) tissues. Shown are the normalized log10 unique molecular identifier (UMI) counts which were averaged across all spots for each gene across both replicates. b, d, H&E images of independent MOB tissue sections which were placed onto separate gasket chambers of the same Array-seq slide. c, e, Unsupervised clustering highlights the histological layers of the MOB tissue using the Leiden algorithm (c, showing clusters annotated by tissue subregions) or indicated algorithms for comparative analyses (e, showing raw clustering results across methods). ONL, Olfactory Nerve Layer; GL, Glomerular Layer; EPL, External Plexiform Layer; MCL, Mitral Cell Layer; IPL, Internal Plexiform Layer; GCL, Granule Cell Layer; RMS, Rostral Migratory Stream. f, Spatial plots of indicated MOB tissue subregions (left panels) and scaled log10 expression of subregion-specific marker genes (right panels) overlaid on grayscale H&E images. Scale bars, 500 µm.
Extended Data Fig. 3 mRNA diffusion and spatial cell type assignment analyses.
a, Images highlighting the spots of the Array-seq slide that were under (orange) or outside (blue) of the two MOB sections which were profiled. b, Spatial plots of total UMIs detected per spot across both MOB Array-seq data sets. c, Density plots (smoothed by kernel density estimation) of the distributions of UMIs per spot detected under (orange) and outside (blue) of the tissue sections. d, Cell type assignments across spots on the Array-seq slide for the indicated MOB cell types (colors) using indicated algorithms (top). Cell types with the highest percentage in inferred proportion per spot were assigned to each spot. EPL-IN, external plexiform layer interneuron; GC, granule cell; M/TC, mitral and tufted cell; PGC, periglomerular cell. Scale bars, 500 µm.
Extended Data Fig. 4 Extended comparison of Array-seq and Visium mouse kidney spatial transcriptomics data.
a, Representative images of virtually rendered Visium and Array-seq spot coverage (18.8% for Visium vs 60.1% for Array-seq). Scale bars, 50 μm. b, c, Bar plots of the number of spots per mm2 (b) and total active area (c) on indicated platforms. d, Downsampling analysis showing changes in sequencing saturation (left) and total genes detected across the entire section (right) using kidney section data from Array-seq (dark gray) and Visium (light gray) platforms (n = 4 per platform). e–h, Bar plots of the numbers of spots under kidney tissue sections (e), total genes and UMIs detected (f), median genes and UMIs detected per spot (g), and genes and UMIs detected per µm2 of tissue on top of the capture area (h). Numbers in e–h were calculated using data that were downsampled to similar levels of sequencing depth. Data are presented as mean values ± SD (n = 4 per platform). i, H&E images (top) overlaid with annotations of tissue subregions (middle) and cell type assignments to spots (bottom) across replicates and platforms (columns). Subregions: CT, Connecting tubule; DCT, Distal Convoluted Tubule; G, Glomerulus; PCT, Proximal Convoluted Tubule; ISOM, Inner Stripe of Outer Medulla; CD, Collecting Duct; OSOM, Outer Stripe of Outer Medulla. Cell types: ATL, thin ascending limb of loop of Henle; CNT, connecting tubule; CTAL, thick ascending limb of loop of Henle in cortex; DCT, distal convoluted tubule; DTL, descending limb of loop of Henle; EC, endothelial cells; ICA, type A intercalated cells of collecting duct; ICB, type B intercalated cells of collecting duct; MTAL, thick ascending limb of loop of Henle in medulla; PC1 and 2; principle cells; PEC, parietal epithelial cells; Per, pericytes; Pod, podocytes; PTS1 and 3, S1 and S3 segments of proximal tubule; Uro, urothelium. Scale bars, 1 mm. j, Bar plots of the proportions of cell type-annotated spots for each to tissue subregion (bottom axis label) in Array-seq and Visium (top axis label; A =Array-seq, V = Visium) kidney tissue sections (n = 1, section pair 1). k, Bar plots of the proportion of spots under a tissue section which matched indicated cell types. Each spot was annotated with the most abundant cell type inferred computationally. Cell types commonly found are in the left panel and rare cell types are in the right panel (black boarder, Array-seq; gray boarder, Visium). Data are presented as mean values ± SD (n = 4 per platform).
Extended Data Fig. 5 Spatial marker gene expression analyses in mouse kidney sections.
a, Scaled log10 expression of indicated marker genes overlaid on grayscale H&E images of matching kidney sections for indicated tissue subregions and platforms (columns). PCT, Proximal Convoluted Tubule; G, Glomerulus; DCT, Distal Convoluted Tubule; ISOM, Inner Stripe of Outer Medulla; ISOM, Inner Stripe of Outer Medulla. Scale bars: 1 mm. b, Heatmaps of differentially expressed genes (rows) for spots (columns) corresponding to indicated kidney tissue subregions (top). Shown are the top five DE genes obtained with Array-seq data and plotted for both Array-seq (left) and Visium (right) datasets. Values are z-scores of log10 normalized UMI counts. c, Correlation (Pearson’s coefficient) between Array-seq and Visium kidney gene expression and whole-kidney, bulk RNA-seq data. Shown are the normalized log10 UMI counts which were averaged across all spots and replicates (n = 4) for indicated spatial platform (Y axis) or across independent replicates (n = 4) for bulk, whole-tissue RNA-seq (X axis).
Extended Data Fig. 6 Three-dimensional Array-seq analysis of serial mouse kidney sections.
a, Images Array-seq data from eight mouse kidney sections aligned in a Z-stack and colored according to z positions within the stack. 80-120 µm were skipped in between each section. b–d, Uniform Manifold Approximation and Projection (UMAP) plots of all spots from Array-seq profiles aggregating all eight serial kidney sections and colored by z position (b), Leiden clusters (c), or manually annotated clusters matching kidney tissue subregions (d). CT, Connecting tubule; DCT, Distal Convoluted Tubule; G, Glomerulus; ISOM, Inner Stripe of Outer Medulla; ISOM, Inner Stripe of Outer Medulla; PCT, Proximal Convoluted Tubule. e, Bar plots of the proportion of spots annotated as belonging to indicated kidney tissue subregions (y axis) for each tissue section (x axis). f, Spatial plots of indicated kidney tissue subregions (leftmost panels) and subregion marker genes (scaled log10 expression) overlaid on grayscale H&E images. Consecutive kidney sections are shown from top (z = 1) to bottom (z = 8). Scale bars, 1 mm.
Extended Data Fig. 7 Reproducibility of Array-seq for multi-organ section profiling.
a, b, Correlation (Pearson’s coefficient) between replicate Array-seq profiles (a), or between average Array-seq and bulk RNA-seq datasets (b) for indicated mouse tissue types. In a, shown are the normalized log10 unique molecular identifier (UMI) counts which were averaged across all spots for each gene across each replicate. In b, Shown are the normalized log10 unique molecular identifier (UMI) counts which were averaged across all spots for Array-seq data (n = 2 for brain and 3 for liver and kidney sections) (y axis) or across independent organ samples (n = 4) for bulk, whole-tissue RNA-seq data (x axis). c, Bar plots of the proportion of the total Array-seq spots under tissue sections which matched indicated tissue subregions. Bars (x axis), replicate sections for each organ type. For brain: Gran. Layer, Granular Layer; Mol. Layer, Molecular Layer. For kidney: DCT, Distal Convoluted Tubule; G, Glomerulus; PCT, Proximal Convoluted Tubule; ISOM, Inner Stripe of Outer Medulla; ISOM, Inner Stripe of Outer Medulla. d, Spatial plots of indicated tissue subregions (leftmost panels) and subregion marker genes (scaled log10 expression) overlaid on grayscale H&E images. Scale bars, 2 mm.
Extended Data Fig. 8 Spatial enrichment of gene ontology gene sets in Array-seq profiles.
a, c, e, Spatial plots of indicated tissue subregions (left) and normalized enrichment score of indicated gene sets (right) in representative kidney (a), brain (c), and liver (e) sections. For brain: Gran. L., Granular Layer; Mol. L., Molecular Layer; Dent., dentate. For kidney: DCT, Distal Convoluted Tubule; G, Glomerulus; PCT, Proximal Convoluted Tubule; ISOM, Inner Stripe of Outer Medulla; ISOM, Inner Stripe of Outer Medulla. Scale bars, 2 mm. b, d, f, Heatmap of enriched Gene Ontology (GO) terms (rows) in indicated tissue subregions (columns) in representative kidney (b), brain (d), and liver (f) sections. Values are row normalized enrichment scores.
Extended Data Fig. 9 Array-seq analysis of a whole-mount, human spleen section.
a, H&E image of a whole-mount, human spleen section mounted onto an Array-seq slide. b–f, Spatial plots of total unique molecular identifiers (UMIs) per spot (b), unsupervised, Leiden clustering results (c), and scaled gene expression for indicated marker genes for B cells (d), macrophages (e), and T cells (f). Scale bars, 5 mm.
Extended Data Fig. 10 Comparison between Array-seq and sequencing-based, spatial transcriptomics methods.
a, Dot plot showing the total surface area available for spatial profiling (y axis) and the diameter of the barcoded spots, beads, or DNA species arranged on the slides or substrates used for mRNA capture (x axis) across indicated spatial transcriptomics methods compatible with fresh-frozen histological sections. Pink dots indicate that a method is compatible with H&E imaging on the same section that is used for spatial profiling. Easy-to-adopt indicates methods which can be readily deployed without the need for special expertise, instrumentation, or custom-made reagents. b, Diagram of Array-seq (left) and Visium (right) slides showing the mRNA capture area (grey) sizes and positions at scale. c–e, Bar plots of the total surface area available for mRNA capture (c), the sensitivity computed in total unique molecular identifiers (UMIs) detected per µm2 (d), and the cost per mm2 of active surface area (e) for indicated method (x axis). In d, the sensitivity analysis was performed using publicly available, preprocessed datasets for each method on MOB tissue, except for Seq-Scope (mouse liver) and DBiT-seq (mouse embryo). In e, asterisks indicate that library preparation costs are included.
Supplementary information
Supplementary Information
Supplementary Table 1: Links to the datasets used for the comparative sensitivity analysis across existing ST methods based on mRNA capture followed by sequencing. Supplementary Table 2: Cost breakdown for Array-seq reagents needed from the initial preparation of Array-seq slides using custom-sequence microarrays to the downstream sequencing library preparation.
Source data
Source Data Fig. 1
Unprocessed gel image for Fig. 1c.
Source Data Extended Data Fig. 1
Statistical source data for Extended Data Fig. 1b,c.
Source Data Extended Data Fig. 4
Statistical source data for Extended Data Fig. 4e–h,k.
Source Data Extended Data Fig. 5
Statistical source data for Extended Data Fig. 5b.
Source Data Extended Data Fig./Table 10
Source values for Extended Data Fig. 10a,c–e.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cipurko, D., Ueda, T., Mei, L. et al. Repurposing large-format microarrays for scalable spatial transcriptomics. Nat Methods 22, 145–155 (2025). https://doi.org/10.1038/s41592-024-02501-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-024-02501-5
This article is cited by
-
Bridging the dimensional gap from planar spatial transcriptomics to 3D cell atlases
Nature Methods (2025)