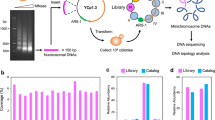
Abstract
The ideal technology for directly investigating the relationship between genotype and phenotype would analyze both RNA and DNA genome-wide and with single-cell resolution; however, existing tools lack the throughput required for comprehensive analysis of complex tumors and tissues. We introduce a highly scalable method for jointly profiling DNA and expression following nucleosome depletion (DEFND-seq). In DEFND-seq, nuclei are nucleosome-depleted, tagmented and separated into individual droplets for messenger RNA and genomic DNA barcoding. Once nuclei have been depleted of nucleosomes, subsequent steps can be performed using the widely available 10x Genomics droplet microfluidic technology and commercial kits. We demonstrate the production of high-complexity mRNA and gDNA sequencing libraries from thousands of individual nuclei from cell lines, fresh and archived surgical specimens for associating gene expression with both copy number and single-nucleotide variants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The sequencing data and count matrices reported in this paper are available in the Gene Expression Omnibus (GSE224149). Fragment files for GBM tumors are stored on Zenodo at https://doi.org/10.5281/zenodo.13984672 (ref. 75). The COSMIC database (https://cancer.sanger.ac.uk/cosmic) was leveraged for SNP and SNV calling. Data from the sci-L3 assay were downloaded from the Sequence Read Archive project (PRJNA511715). Bulk BJ WGS data were acquired from GSE63577 (SRR1660537) and 10x ATAC PBMC data were acquired from 10x Genomics17. Source data are provided with this paper.
Code availability
All source code is available on the Sims Laboratory GitHub repository at https://github.com/simslab/, including code for generating expression count matrices (DropSeqPipeline8), clustering and differential expression (cluster_diffex2018) and genomic DNA analysis (dna10x).
References
Kashima, Y. et al. Single-cell sequencing techniques from individual to multiomics analyses. Exp. Mol. Med. 52, 1419–1427 (2020).
Mallory, X. F., Edrisi, M., Navin, N. & Nakhleh, L. Methods for copy number aberration detection from single-cell DNA-sequencing data. Genome Biol. 21, 1–22 (2020).
Suvà, M. L. & Tirosh, I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol. Cell 75, 7–12 (2019).
Macaulay, I. C., Ponting, C. P. & Voet, T. Single-cell multiomics: multiple measurements from single cells. Trends Genet. 33, 155–168 (2017).
Darmanis, S. et al. Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 21, 1399–1410 (2017).
Lawson, D. A., Kessenbrock, K., Davis, R. T., Pervolarakis, N. & Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 20, 1349–1360 (2018).
Dey, S. S., Kester, L., Spanjaard, B., Bienko, M. & van Oudenaarden, A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 33, 285–289 (2015).
Macaulay, I. C. et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522 (2015).
Reuter, J. A., Spacek, D. V., Pai, R. K. & Snyder, M. P. Simul-seq: combined DNA and RNA sequencing for whole-genome and transcriptome profiling. Nat. Methods 13, 953–958 (2016).
Han, K. Y. et al. SIDR: simultaneous isolation and parallel sequencing of genomic DNA and total RNA from single cells. Genome Res. 28, 75–87 (2018).
Nam, A. S. et al. Somatic mutations and cell identity linked by genotyping of transcriptomes. Nature 571, 355–360 (2019).
Miles, L. A. et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 587, 477–482 (2020).
Yin, Y. et al. High-throughput single-cell sequencing with linear amplification. Mol. Cell 76, 676–690. e610 (2019).
Vitak, S. A. et al. Sequencing thousands of single-cell genomes with combinatorial indexing. Nat. Methods 14, 302–308 (2017).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Chen, C. et al. Single-cell whole-genome analyses by linear amplification via transposon insertion (LIANTI). Science 356, 189–194 (2017).
10x Genomics. 500 peripheral blood mononuclear cells (PBMCs) from a healthy donor (Next GEM v.1.1) (2021); https://www.10xgenomics.com/datasets/500-peripheral-blood-mononuclear-cells-pbm-cs-from-a-healthy-donor-next-gem-v-1-1-1-1-standard-2-0-0
Gonzalez-Pena, V. et al. Accurate genomic variant detection in single cells with primary template-directed amplification. Proc. Natl Acad. Sci. USA 118, e2024176118 (2021).
Marthandan, S. et al. Conserved senescence associated genes and pathways in primary human fibroblasts detected by RNA-Seq. PLoS ONE 11, e0154531 (2016).
Davis, M. E. Glioblastoma: overview of disease and treatment. Clin. J. Oncol. Nurs. 20, S2 (2016).
Rohle, D. et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 340, 626–630 (2013).
Wang, J. et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 48, 768–776 (2016).
Verhaak, R. G. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer Cell 17, 98–110 (2010).
Yuan, J. et al. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 10, 1–15 (2018).
Chaligne, R. et al. Epigenetic encoding, heritability and plasticity of glioma transcriptional cell states. Nat. Genet. 53, 1469–1479 (2021).
Neftel, C. et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849.e821 (2019).
Zhao, W. et al. Deconvolution of cell type-specific drug responses in human tumor tissue with single-cell RNA-seq. Genome Med. 13, 82 (2021).
Reifenberger, G., Liu, L., Ichimura, K., Schmidt, E. E. & Collins, V. P. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res. 53, 2736–2739 (1993).
Lei, L. et al. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS ONE 6, e20041 (2011).
Szerlip, N. J. et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl Acad. Sci. USA 109, 3041–3046 (2012).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
Krishna, K. V., Dubey, S. K., Singhvi, G., Gupta, G. & Kesharwani, P. MAPK pathway: potential role in glioblastoma multiforme. Interdiscip. Neurosurg. 23, 100901 (2021).
Hui, A. B.-Y., Lo, K.-W., Yin, X.-L., Poon, W.-S. & Ng, H.-K. Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab. Invest. 81, 717–723 (2001).
Arslan, S. et al. Sequencing by avidity enables high accuracy with low reagent consumption. Nat. Biotechnol. 42, 132–138 (2024).
Tate, J. G. et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019).
Nickel, G. C. et al. Characterizing mutational heterogeneity in a glioblastoma patient with double recurrence. PLoS ONE 7, e35262 (2012).
De Souza, C. et al. Effect of the p53 P72R polymorphism on mutant TP53 allele selection in human cancer. J. Natl Cancer Inst. 113, 1246–1257 (2021).
Kang, H. M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36, 89–94 (2018).
McFarland, J. M. et al. Multiplexed single-cell transcriptional response profiling to define cancer vulnerabilities and therapeutic mechanism of action. Nat. Commun. 11, 4296 (2020).
Gaublomme, J. T. et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat. Commun. 10, 2907 (2019).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e289 (2019).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Rodriguez-Meira, A. et al. Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol. Cell 73, 1292–1305.e1298 (2019).
Datlinger, P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e1817 (2016).
Raj, B. et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 36, 442–450 (2018).
Yang, D. et al. Lineage tracing reveals the phylodynamics, plasticity, and paths of tumor evolution. Cell 185, 1905–1923.e1925 (2022).
Biddy, B. A. et al. Single-cell mapping of lineage and identity in direct reprogramming. Nature 564, 219–224 (2018).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Al-Dalahmah, O. et al. Single-nucleus RNA-seq identifies Huntington disease astrocyte states. Acta Neuropathol. Commun. 8, 1–21 (2020).
Krishnaswami, S. R. et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc. 11, 499–524 (2016).
Hobson, B. D. et al. Subcellular and regional localization of mRNA translation in midbrain dopamine neurons. Cell Rep. 38, 110208 (2022).
Snyder, M. E. et al. Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci. Immunol. 4, eaav5581 (2019).
Yuan, J. & Sims, P. A. An automated microwell platform for large-scale single cell RNA-seq. Sci. Rep. 6, 33883 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Levitin, H. M. et al. De novo gene signature identification from single-cell RNA-seq with hierarchical Poisson factorization. Mol. Syst. Biol. 15, e8557 (2019).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
McInnes, L., Healy, J. & Melville, J. UMAP: Uniform Manifold Approximation and Projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
Shekhar, K. et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323. e1330 (2016).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Fang, R. et al. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 12, 1337 (2021).
Zhang, K. et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 184, 5985–6001.e5919 (2021).
Lai, D. shahcompbio/HMMcopy_utils. GitHub https://github.com/shahcompbio/hmmcopy_utils (2011).
Shah, S. P. et al. Integrating copy number polymorphisms into array CGH analysis using a robust HMM. Bioinformatics 22, e431–e439 (2006).
Ha, G. et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 22, 1995–2007 (2012).
Zahn, H. et al. Scalable whole-genome single-cell library preparation without preamplification. Nat. Methods 14, 167–173 (2017).
Van der Auwera, G. A. & O’Connor, B. D. Genomics in the Cloud: using Docker, GATK, and WDL in Terra (O’Reilly Media, 2020).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164–e164 (2010).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Yu, L. et al. scONE-seq: a single-cell multi-omics method enables simultaneous dissection of phenotype and genotype heterogeneity from frozen tumors. Sci. Adv. 9, eabp8901 (2023).
Li, H. lh3/seqtk. GitHub https://github.com/lh3/seqtk (2012).
Olsen, T. Processed GBM DEFND-seq data. Zenodo https://doi.org/10.5281/zenodo.13984672 (2024).
Acknowledgements
Some of the research reported here was performed in the Sulzberger Columbia Genome Center, which is supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) grant P30CA013696 and NIH/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant P30DK132710. P.A.S. and S.Z. were supported by a RISE grant from Columbia University. P.A.S. was supported by NIH/NCI grant U54CA274506. P.A.S., P.C., and J.N.B. were supported by NIH/National Institute of Neurological Disorders and Stroke (NINDS) grant R01NS103473. S.Z. was supported by NIH/NCI grant R01CA275184. P.T. was supported by the I.I. Rabi Scholars Program of Columbia University.
Author information
Authors and Affiliations
Contributions
T.R.O., S.Z. and P.A.S. conceived experiments. T.R.O., P.T., R.K.S. and P.A.S. analyzed data. T.R.O. and P.T. performed the experiments. J.F. managed biobanked samples. P.C. and J.N.B. procured and characterized specimens. T.R.O. and P.A.S. wrote the manuscript. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.A.S. receives patent royalties from Guardant Health. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Lei Tang and Hui Hua, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Copy number variation heatmap of freshly resected GBM tumor cells. Cells are grouped by gDNA cluster.
Chromosome location reference on bottom margin.
Extended Data Fig. 2 DEFND-seq applied to cryopreserved GBM tumor sample.
(a) Clustering of GBM cells by gene expression. (b) Same as (a) but colored by expression of MOBP. (c) Highly expressed genes for each cluster in (a). (d) log fold change of PTEN in the genome of the clusters in (a) showing deletions in clusters 1–6. Bars indicate the medians of each cluster. (e) log fold change of MYCN in the genome of clusters in (a) with amplifications in clusters 1–6. (f) Same as (e) but with CDK4. (g) – (i) Cells from (a) colored by genomic log fold change in (g) PTEN, (h) MYCN, and (i) CDK4. (j) – (l) Cells from (a) colored by log-normalized transcript counts for (j) PTEN, (k) MYCN, and (l) CDK4. (m) Pileup of every base along selected genomic regions for all transformed cells. PTEN, MYCN, and CDK4 regions are presented along with a selection of genes located in the sampled window.
Extended Data Fig. 3 Copy number variation heatmap of cryopreserved GBM tumor cells.
Cells are grouped by gene expression cluster. Chromosome location reference on bottom margin.
Extended Data Fig. 4 SNP and SNV detection.
(a) Sequencing saturation analysis of the number of unique fragments per nucleus for a GBM DEFND-seq library sequenced on an Illumina NovaSeq 6000, and on an Element Aviti. (b) Sequencing saturation analysis of the transcription start site enrichment for a library sequenced on both NovaSeq and Aviti sequencers. (c) Insert size distribution for the GBM library sequenced on NovaSeq and Aviti sequencers. (d) PREX1 genomic mutation detection projected on a UMAP derived from the gene expression dataset. (e) Same as (d) but colored by PREX1 expression. (f) Average frequency of PREX1 variant allele and average expression of PREX1 in astrocyte-like, proliferating, and proneural clusters. (g) Integrated genomic viewer (IGV) style plots of the transformed cells showing exonic regions of PREX1, coverage, and the location of the SNV49. The SNV is magnified to show the missense mutation and affected amino acid. Individual reads are shown for both the NovaSeq and Aviti sequencers and each read is colored by cell of origin (cell barcode). (h) Same as (g) but for the p.P72R mutation on TP53.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Tables 1 and 2 and Protocol.
Source data
Source Data Fig. 1
Statistical Source Data for Fig. 1.
Source Data Fig. 2
Statistical Source Data for Fig. 2.
Source Data Fig. 3
Statistical Source Data (individual data points for Fig. 3).
Source Data Fig. 4
Statistical Source Data for Fig. 4.
Source Data Fig. 5
Statistical Source Data (.xlsx) for Fig. 5.
Source Data Fig. 6
Statistical Source Data for Fig. 6.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Olsen, T.R., Talla, P., Sagatelian, R.K. et al. Scalable co-sequencing of RNA and DNA from individual nuclei. Nat Methods 22, 477–487 (2025). https://doi.org/10.1038/s41592-024-02579-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-024-02579-x
This article is cited by
-
Year in review 2025
Nature Methods (2026)
-
Functional phenotyping of genomic variants using joint multiomic single-cell DNA–RNA sequencing
Nature Methods (2025)
-
Simultaneous single-cell sequencing of RNA and DNA at scale with DEFND-seq
Nature Reviews Genetics (2025)
-
Engineering next-generation microfluidic technologies for single-cell phenomics
Nature Genetics (2025)
-
Precisely defining disease variant effects in CRISPR-edited single cells
Nature (2025)