Abstract
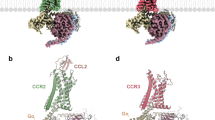
Chemokines regulate immune cell migration in development, homeostasis and inflammation, but the precise spatiotemporal pattern of chemokine release in vivo remains elusive due to the constraints of existing detection methodologies. Here, we report the engineering and characterization of a genetically encoded green fluorescent chemokine sensor, named CRAFi-CCR2, which utilizes the CCR2 receptor as a sensing moiety. In astrocytes, hCRAFi-CCR2, derived from the human CCR2B receptor, exhibited ~300% increase in fluorescence in response to mCCL2, with nanomolar affinity (2.5 nM). Activation of hCRAFi-CCR2 did not affect downstream signaling pathways, such as calcium mobilization and receptor internalization. Using this sensor, we performed 17–20 h of real-time imaging to observe endogenous mCCL2 release under inflammatory conditions, both in cell culture and in mice. In mouse brain, we observed spatial heterogeneity of CCL2 signal response on a scale of about 20–50 µm, highlighting the complexity of the immune system’s spatiotemporal signaling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All essential raw datasets, including files for supplementary figures, the mass spectrometry proteomics data including raw datasets, and raw unprocessed images, are available at figshare via https://figshare.com/projects/CRAFi-CCR2/242891 (ref. 63). The rest of the files are available from the corresponding author upon request, as the total size of the files acquired for this study exceeds the 20-GB limit of the figshare repository. All plasmids used in this study are available from WeKwikGene (https://wekwikgene.wllsb.edu.cn/publications/1173; accession number 0001173-0001182) and Addgene. Source data are provided with this paper.
Code availability
Custom MATLAB codes used for data acquisition are available at https://github.com/MingCShare/xyzRegist/.
References
Hughes, C. E. & Nibbs, R. J. B. A guide to chemokines and their receptors. FEBS J. 285, 2944–2971 (2018).
Sokol, C. L. & Luster, A. D. The chemokine system in innate immunity. Cold Spring Harb. Perspect. Biol. 7, a016303 (2015).
Proudfoot, AmandaE. I. Chemokine receptors: multifaceted therapeutic targets. Nat. Rev. Immunol. 2, 106–115 (2002).
Dinarello, C. A. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 27, 519–550 (2009).
Bradley, J. R. TNF-mediated inflammatory disease. J. Pathol. 214, 149–160 (2008).
Bours, M. J. L., Swennen, E. L. R., Di Virgilio, F., Cronstein, B. N. & Dagnelie, P. C. Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol. Ther. 112, 358–404 (2006).
Tang, P. & Wang, J. M. Chemokines: the past, the present and the future. Cell. Mol. Immunol. 15, 295–298 (2018).
Duan, L. et al. PDGFRβ cells rapidly relay inflammatory signal from the circulatory system to neurons via chemokine CCL2. Neuron 100, 183–200 (2018).
Van Steenwinckel, J. et al. CCL2 released from neuronal synaptic vesicles in the spinal cord is a major mediator of local inflammation and pain after peripheral nerve injury. J. Neurosci. 31, 5865–5875 (2011).
Mahajan, S. D., Schwartz, S. A. & Nair, M. P. N. Immunological assays for chemokine detection in In-vitro culture of CNS cells. Biol. Proced. Online 5, 90–102 (2003).
McDonough, A. A., Veiras, L. C., Minas, J. N. & Ralph, D. L. Considerations when quantitating protein abundance by immunoblot. Am. J. Physiol. Cell Physiol. 308, C426–C433 (2015).
Maubert, M. A. et al. High-resolution mass spectrometry and partial de novo sequencing constitute a useful approach for determining the profile of chemokine secretion following the stimulation of human intestinal epithelial cells. Rapid Commun. Mass Spectrom. 27, 2179–2187 (2013).
Trenchevska, O. et al. Quantitative mass spectrometric immunoassay for the chemokine RANTES and its variants. J. Proteom. 116, 15–23 (2015).
Sánchez-Tirado, E., Agüí, L., González-Cortés, A., Yáñez-Sedeño, P. & Pingarrón, J. M. Biodetection techniques for quantification of chemokines. Chemosensors 10, 294 (2022).
Aydın, E. B. & Sezgintürk, M. K. Fabrication of electrochemical immunosensor for detection of interleukin 8 biomarker via layer-by-layer self-assembly process on cost-effective fluorine tin oxide electrode. Electroanalysis 33, 1596–1605 (2021).
White, C. W., Kilpatrick, L. E., Pfleger, K. D. G. & Hill, S. J. A nanoluciferase biosensor to investigate endogenous chemokine secretion and receptor binding. iScience 24, 102011 (2021).
Wu, Z., Lin, D. & Li, Y. Pushing the frontiers: tools for monitoring neurotransmitters and neuromodulators. Nat. Rev. Neurosci. 23, 257–274 (2022).
Singh, S., Anshita, D. & Ravichandiran, V. MCP-1: function, regulation, and involvement in disease. Int. Immunopharmacol. 101, 107598 (2021).
Charo, I. F. et al. Molecular cloning and functional expression of two monocyte chemoattractant protein 1 receptors reveals alternative splicing of the carboxyl-terminal tails. Proc. Natl Acad. Sci. USA 91, 2752–2756 (1994).
Wong, L. M. et al. Organization and differential expression of the human monocyte chemoattractant protein 1 receptor gene: evidence for the role of the carboxyl-terminal tail in receptor trafficking. J. Biol. Chem. 272, 1038–1045 (1997).
Piatkevich, K. D. et al. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 14, 352–360 (2018).
Hall, S. E. et al. Elucidation of binding sites of dual antagonists in the human chemokine receptors CCR2 and CCR5. Mol. Pharmacol. 75, 1325–1336 (2009).
Weber, M. et al. Interstitial dendritic cell guidance by haptotactic chemokine gradients. Science 339, 328–332 (2013).
Berchiche, Y. A., Gravel, S. P., Pelletier, M. E., St-Onge, G. & Heveker, N. Different effects of the different natural CC-chemokine receptor 2B (CCR2B) ligands on β-arrestin recruitment, Gαi signalling, and receptor internalization. Mol. Pharmacol. 79, 488–498 (2010).
Huma, Z. E. et al. Key determinants of selective binding and activation by the monocyte chemoattractant proteins at the chemokine receptor CCR2. Sci. Signal. 10, eaai8529 (2017).
Islam, S. A. et al. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ TH2 cells. Nat. Immunol. 12, 167–177 (2011).
Chui, R. & Dorovini-Zis, K. Regulation of CCL2 and CCL3 expression in human brain endothelial cells by cytokines and lipopolysaccharide. J. Neuroinflammation 7, 1 (2010).
Thompson, W. L. & Van Eldik, L. J. Inflammatory cytokines stimulate the chemokines CCL2/MCP-1 and CCL7/MCP-7 through NFκB and MAPK dependent pathways in rat astrocytes. Brain Res. 1287, 47–57 (2009).
Sierra-Filardi, E. et al. CCL2 shapes macrophage polarization by GM-CSF and M-CSF: identification of CCL2/CCR2-dependent gene expression profile. J. Immunol. 192, 3858–3867 (2014).
Cerri, C. et al. The chemokine CCL2 mediates the seizure-enhancing effects of systemic inflammation. J. Neurosci. 36, 3777–3788 (2016).
Liu, J. et al. Screening cytokine/chemokine profiles in serum and organs from an endotoxic shock mouse model by LiquiChip. Sci. China Life Sci. 60, 1242–1250 (2017).
Renner, N. A., Ivey, N. S., Redmann, R. K., Lackner, A. A. & MacLean, A. G. MCP-3/CCL7 production by astrocytes: implications for SIV neuroinvasion and AIDS encephalitis. J. Neurovirol. 17, 146–152 (2011).
Orchanian, S. B., Still, K., Harris, T. H. & Lodoen, M. B. Deficiency in astrocyte CCL2 production reduces neuroimmune control of Toxoplasma gondii infection. PLoS Pathog. 20, e1011710 (2024).
Hasel, P., Rose, I. V. L., Sadick, J. S., Kim, R. D. & Liddelow, S. A. Neuroinflammatory astrocyte subtypes in the mouse brain. Nat. Neurosci. 24, 1475–1487 (2021).
Carpentier, P. A. et al. Differential activation of astrocytes by innate and adaptive immune stimuli. Glia 49, 360–374 (2005).
Rodgers, K. R., Lin, Y., Langan, T. J., Iwakura, Y. & Chou, R. C. Innate immune functions of astrocytes are dependent upon tumor necrosis factor-alpha. Sci. Rep. 10, 7047 (2020).
Zhou, Y., Ling, E. A. & Dheen, S. T. Dexamethasone suppresses monocyte chemoattractant protein-1 production via mitogen activated protein kinase phosphatase-1 dependent inhibition of Jun N-terminal kinase and p38 mitogen-activated protein kinase in activated rat microglia. J. Neurochem. 102, 667–678 (2007).
Lu, Y. C., Yeh, W. C. & Ohashi, P. S. LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151 (2008).
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
Liu, S. et al. Hyperbaric oxygen alleviates the inflammatory response induced by LPS through inhibition of NF-κB/MAPKs-CCL2/CXCL1 signaling pathway in cultured astrocytes. Inflammation 41, 2003–2011 (2018).
Erickson, M. A. & Banks, W. A. Cytokine and chemokine responses in serum and brain after single and repeated injections of lipopolysaccharide: multiplex quantification with path analysis. Brain. Behav. Immun. 25, 1637–1648 (2011).
Semple, B. D., Bye, N., Rancan, M., Ziebell, J. M. & Morganti-Kossmann, M. C. Role of CCL2 (MCP-1) in traumatic brain injury (TBI): evidence from severe TBI patients and CCL2−/− mice. J. Cereb. Blood Flow. Metab. 30, 769–782 (2010).
Chen, Y. et al. Spatiotemporally selective astrocytic ATP dynamics encode injury information sensed by microglia following brain injury in mice. Nat. Neurosci. 27, 1522–1533 (2024).
Su, L., Shen, G. & Sun, L. Role of CXCL1 and CCL2 in patients with severe traumatic brain injury. Ann. Phys. Rehabil. Med. 61, e231 (2018).
Feng, J. et al. A genetically encoded fluorescent sensor for rapid and specific in vivo detection of norepinephrine. Neuron 102, 745–761 (2019).
Ulvmar, M. H. et al. The atypical chemokine receptor CCRL1 shapes functional CCL21 gradients in lymph nodes. Nat. Immunol. 15, 623–630 (2014).
Vaahtomeri, K., Moussion, C., Hauschild, R. & Sixt, M. Shape and function of interstitial chemokine CCL21 gradients are independent of heparan sulfates produced by lymphatic endothelium. Front. Immunol. 12, 630002 (2021).
Batiuk, M. Y. et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 11, 1220 (2020).
Panenka, W. et al. P2X7-like receptor activation in astrocytes increases chemokine monocyte chemoattractant protein-1 expression via mitogen-activated protein kinase. J. Neurosci. 21, 7135–7142 (2001).
Shieh, C. H., Heinrich, A., Serchov, T., van Calker, D. & Biber, K. P2X7-dependent, but differentially regulated release of IL-6, CCL2, and TNF-α in cultured mouse microglia. Glia 62, 592–607 (2014).
Villette, V. et al. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell 179, 1590–1608 (2019).
Zhang, H. et al. Quantitative assessment of near-infrared fluorescent proteins. Nat. Methods 20, 1605–1616 (2023).
Dou, Y. et al. Microglial migration mediated by ATP-induced ATP release from lysosomes. Cell Res. 22, 1022–1033 (2012).
Hughes, C. S. et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 14, 68–85 (2018).
Wu, Z. et al. A sensitive GRAB sensor for detecting extracellular ATP in vitro and in vivo. Neuron 110, 770–782 (2022).
Lagraoui, M. et al. Controlled cortical impact and craniotomy induce strikingly similar profiles of inflammatory gene expression, but with distinct kinetics. Front. Neurol. 3, 155 (2012).
Xu, J. et al. Astrocyte-derived CCL2 participates in surgery-induced cognitive dysfunction and neuroinflammation via evoking microglia activation. Behav. Brain Res. 332, 145–153 (2017).
Davalos, D. et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8, 752–758 (2005).
Raduolovic, K., Mak’Anyengo, R., Kaya, B., Steinert, A. & Niess, J. H. Injections of lipopolysaccharide into mice to mimic entrance of microbial-derived products after intestinal barrier breach. J. Vis. Exp. 57610 (2018).
Lubkowski, J. et al. The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions. Nat. Struct. Biol. 4, 64–69 (1997).
Oberthür, D. et al. Crystal structure of a mirror-image l-RNA aptamer (Spiegelmer) in complex with the natural l-protein target CCL2. Nat. Commun. 6, 6923 (2015).
Rodríguez-Frade, J. M. et al. The chemokine monocyte chemoattractant protein-1 induces functional responses through dimerization of its receptor CCR2. Proc. Natl Acad. Sci. USA 96, 3628–3633 (1999).
Xiao, X. et al. Data source files for the manuscript "Genetically Encoded Biosensor for Monitoring Spatiotemporal Dynamics of CCR2 Ligands in Culture and In Vivo". figshare https://figshare.com/projects/CRAFi-CCR2/242891 (2025).
Acknowledgements
We thank F. Li, S. Papadaki and Y. Y. Han from the laboratory of K.D.P. for their management support; Y. P. Gao, M. H. Liao, F. Xiao and L. M. Zheng, G.C. Fang from the Imaging Core at Westlake University for their technical support with the Nikon and Zeiss confocal microscopes; S. S. Liu from the Core Facilities at Zhejiang University School of Medicine for assistance with the two-photon microscope; F. Yang at Zhejiang University School of Medicine for support in generating the 3D model of hCRAFi-CCR2 sensor using AlphaFold2; M. Z. Fan and J. Hu from the Mass Spectrometry & Metabolomics Core Facility at Westlake University for assistance and discussion with LC–MS/MS analysis; D. Y. Gu from the Instrumentation and Service Center for Molecular Sciences at Westlake University for assistance with spectra measurement; and the Laboratory Animal Resources Center, Biomedical Research Core Facilities, Flow Cytometry Core, Westlake Center for Micro/Nano Fabrication. This work was supported by start-up funding from the Foundation of Westlake University, Westlake Laboratory of Life Sciences and Biomedicine, National Natural Science Foundation of China grants W2432024 and 32171093; and the ‘Pioneer’ and ‘Leading Goose’ R&D Program of Zhejiang 2024SSYS0031 (to K.D.P.), the Young Scientist Program of National Natural Science Foundation of China (grant 32100897 to X.X.), the National Natural Science Foundation of China (32371150 to M.J.) and STI2030-Major project 2021ZD0202200, Subject 2021ZD0202203 (to M.J.).
Author information
Authors and Affiliations
Contributions
X.X. and K.D.P. initiated the project, made high-level designs and plans, and interpreted the data. X.X. developed the sensors with help from G.L. B.S. and X.X. developed the iDrug system. X.X. performed cell culture characterization. X.X. and Q.Q. performed affinity characterization. F.X. performed fast sensor kinetic measurements; X.G., X.S. and X.X. performed immunohistochemistry analysis. C.W., M.J., M.C. and X.X. performed two-photon in vivo experiments and analysis. X.X. and K.D.P., with help from M.J., wrote the manuscript. K.D.P. oversaw all aspects of the project.
Corresponding author
Ethics declarations
Competing interests
B.S. is the founder of CellSorter KFT, which is developing an automated micropipette. X.X. and K.D.P. are inventors on a patent application covering the design and application of the sensors. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Nina Vogt, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–5, Supplementary Tables 1–4 and Supplementary Figs. 1–22.
Supplementary Video 1
Drug-delivery system for PCR mutant library screening. To screen the variants efficiently and economically, we modified the CellSorter from cell picking into a drug-delivery system. During screening, cells were first imaged for 10–20 s to record the baseline fluorescence, and then the manipulator-guided glass pipette was lowered down into the well, 300–400 µm above the cells. Drugs were puffed to the cells through a 70-µm diameter glass capillary connected to the syringe pump, and time-lapse movies were recorded. This video shows how the manipulator-guided glass pipette moves from one well to another well to deliver drugs in an automatic mode.
Supplementary Video 2
Fluorescence response of hCRAFi-CCR2 sensor in HEK cells using the drug-delivery system. Images were acquired at 1 Hz.
Supplementary Video 3
In vivo two-photon imaging of the dynamics of fluorescence intensity for the hCRAFi-CCR2 sensor in mouse visual cortex after LPS challenge. The total time duration is 46 h. Images were acquired at 10-min intervals. Left, raw images showing the hCRAFi-CCR2 fluorescence intensity changes; right, calculated fluorescence response (∆F/F). Lines represent boundaries of the identified signal specific patches.
Supplementary Video 4
In vivo two-photon imaging of the dynamics of fluorescence intensity for hCRAFi-CCR2 sensor or null mutant in mouse visual cortex after laser-induced ablation. Images were acquired at 15-min intervals.
Source data
Source Data Fig. 1
Numerical source data.
Source Data Fig. 2
Numerical source data.
Source Data Fig. 3
Numerical source data.
Source Data Fig. 4
Numerical source data.
Source Data Fig. 5
Numerical source data.
Source Data Fig. 6
Numerical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, X., Wang, C., Guo, X. et al. Genetically encoded biosensor for monitoring spatiotemporal dynamics of CCR2 ligands in culture and in vivo. Nat Methods 22, 1731–1741 (2025). https://doi.org/10.1038/s41592-025-02742-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02742-y