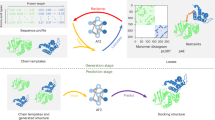
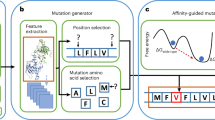
Abstract
Protein complex structure prediction is crucial for understanding of biological activities and advancing drug development. While various experimental methods can provide structural insights into protein complexes, the knowledge obtained is often sparse or approximate. A general tool is needed to integrate limited experimental information for high-throughput and accurate prediction. Here we introduce GRASP to efficiently and flexibly incorporate diverse forms of experimental information. GRASP outperforms existing tools in handling both simulated and real-world experimental restraints including those from crosslinking, covalent labeling, chemical shift perturbation and deep mutational scanning. For example, GRASP excels at predicting antigen–antibody complex structures, even surpassing AlphaFold3 when using experimental deep mutational scanning or covalent-labeling restraints. Beyond its accuracy and flexibility in restrained structure prediction, GRASP’s ability to integrate multiple forms of restraints enables integrative modeling. We also showcase its potential in modeling protein structural interactome under near-cellular conditions using previously reported large-scale in situ crosslinking data for mitochondria.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All relevant evaluation datasets and results are available via OSF at https://doi.org/10.17605/OSF.IO/6KJUQ (ref. 70). Further training data for MEGAFold-Multimer and GRASP are released at http://ftp.cbi.pku.edu.cn/pub/psp/.
Code availability
The GRASP code is available via GitHub at https://github.com/xiergo/GRASP-JAX and via Zenodo at https://doi.org/10.5281/zenodo.15347070 (ref. 71) under the Apache v.2.0 license. Model weights have been deposited on OSF at https://doi.org/10.17605/OSF.IO/6KJUQ (ref. 70). In addition, a Colab notebook is available via GitHub at https://colab.research.google.com/github/xiergo/GRASP-JAX/blob/main/GRASP-JAX.ipynb for ease of use.
References
Pearce, R. & Zhang, Y. Toward the solution of the protein structure prediction problem. J. Biol. Chem. 297, 100870 (2021).
UniProt, Consortium. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2023).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv https://doi.org/10.1101/2021.10.04.463034 (2022).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Wu, R. et al. High-resolution de novo structure prediction from primary sequence. Preprint at bioRxiv https://doi.org/10.1101/2022.07.21.500999 (2022).
Bryant, P. Deep learning for protein complex structure prediction. Curr. Opin. Struct. Biol. 79, 102529 (2023).
Graziadei, A. & Rappsilber, J. Leveraging crosslinking mass spectrometry in structural and cell biology. Structure 30, 37–54 (2022).
Leitner, A., Faini, M., Stengel, F. & Aebersold, R. Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 41, 20–32 (2016).
Vogeli, B. The nuclear Overhauser effect from a quantitative perspective. Prog. Nucl. Magn. Reson. Spectrosc. 78, 1–46 (2014).
Limpikirati, P., Liu, T. & Vachet, R. W. Covalent labeling-mass spectrometry with non-specific reagents for studying protein structure and interactions. Methods 144, 79–93 (2018).
Williamson, M. P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 73, 1–16 (2013).
Nerli, S., McShan, A. C. & Sgourakis, N. G. Chemical shift-based methods in NMR structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 106-107, 1–25 (2018).
Fowler, D. M. & Fields, S. Deep mutational scanning: a new style of protein science. Nat. Methods 11, 801–807 (2014).
Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310.e1220 (2020).
Berger, C. et al. Cryo-electron tomography on focused ion beam lamellae transforms structural cell biology. Nat. Methods 20, 499–511 (2023).
Puthenveetil, R. & Vinogradova, O. Solution NMR: a powerful tool for structural and functional studies of membrane proteins in reconstituted environments. J. Biol. Chem. 294, 15914–15931 (2019).
Chavez, J. D. et al. Systems structural biology measurements by in vivo cross-linking with mass spectrometry. Nat. Protoc. 14, 2318–2343 (2019).
Stahl, K., Graziadei, A., Dau, T., Brock, O. & Rappsilber, J. Protein structure prediction with in-cell photo-crosslinking mass spectrometry and deep learning. Nat. Biotechnol. 41, 1810–1819 (2023).
Stahl, K. et al. Modelling protein complexes with crosslinking mass spectrometry and deep learning. Nat. Commun. 15, 7866 (2024).
Feng, S. et al. Integrated structure prediction of protein–protein docking with experimental restraints using ColabDock. Nat. Mach. Intell. 6, 924–935 (2024).
de Vries, S. J., van Dijk, M. & Bonvin, A. M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 (2010).
Dominguez, C., Boelens, R. & Bonvin, A. M. J. J. HADDOCK: a protein−protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003).
Kozakov, D. et al. The ClusPro web server for protein-protein docking. Nat. Protoc. 12, 255–278 (2017).
Comeau, S. R., Gatchell, D. W., Vajda, S. & Camacho, C. J. ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res. 32, W96–W99 (2004).
Comeau, S. R., Gatchell, D. W., Vajda, S. & Camacho, C. J. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45–50 (2004).
Drake, Z. C., Seffernick, J. T. & Lindert, S. Protein complex prediction using Rosetta, AlphaFold, and mass spectrometry covalent labeling. Nat. Commun. 13, 7846 (2022).
Russel, D. et al. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 10, e1001244 (2012).
Molnar, K. S. et al. Cys-scanning disulfide crosslinking and Bayesian modeling probe the transmembrane signaling mechanism of the histidine kinase, PhoQ. Structure 22, 1239–1251 (2014).
Bayrhuber, M. et al. Structure of the human voltage-dependent anion channel. Proc. Natl Acad. Sci. USA 105, 15370–15375 (2008).
Dauden, M. I. et al. Architecture of the yeast Elongator complex. EMBO Rep. 18, 264–279 (2017).
Sirui, L. et al. Assisting and accelerating NMR assignment with restrained structure prediction. Commun. Biol. 8, 1067 (2025).
Chaudhury, S., Lyskov, S. & Gray, J. J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010).
Alford, R. F. et al. Correction to ‘The Rosetta all-atom energy function for macromolecular modeling and design’. J. Chem. Theory Comput. 18, 4594 (2022).
Lai, J. K., Ambia, J., Wang, Y. & Barth, P. Enhancing structure prediction and design of soluble and membrane proteins with explicit solvent-protein interactions. Structure 25, 1758–1770 e1758 (2017).
Bartolec, T. K. et al. Cross-linking mass spectrometry discovers, evaluates, and corroborates structures and protein-protein interactions in the human cell. Proc. Natl Acad. Sci. USA 120, e2219418120 (2023).
Wang, L., Xie, L. & Zhang, Z. Determination of HER2 binding domain in antigen-antibody complexes based on chemical crosslinking mass spectrometry. J. Proteom. 286, 104954 (2023).
Di Ianni, A., Di Ianni, A., Cowan, K., Barbero, L. M. & Sirtori, F. R. Leveraging cross-linking mass spectrometry for modeling antibody-antigen complexes. J. Proteome Res. 23, 1049–1061 (2024).
Wang, X. et al. Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol. Cell Proteom. 16, 840–854 (2017).
Kalisman, N., Adams, C. M. & Levitt, M. Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc. Natl Acad. Sci. USA 109, 2884–2889 (2012).
Blech, M. et al. One target—two different binding modes: structural insights into gevokizumab and canakinumab interactions to interleukin-1β. J. Mol. Biol. 425, 94–111 (2013).
Vreven, T. et al. Updates to the integrated protein-protein interaction benchmarks: Docking Benchmark Version 5 and Affinity Benchmark Version 2. J. Mol. Biol. 427, 3031–3041 (2015).
Hitawala, F. N. & Gray, J. J. What has AlphaFold3 learned about antibody and nanobody docking, and what remains unsolved? Preprint at bioRxiv https://doi.org/10.1101/2024.09.21.614257 (2024).
Cao, Y. et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 614, 521–529 (2023).
Ruffolo, J. A., Chu, L. S., Mahajan, S. P. & Gray, J. J. Fast, accurate antibody structure prediction from deep learning on massive set of natural antibodies. Nat. Commun. 14, 2389 (2023).
Kosinski, J. et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352, 363–365 (2016).
Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. Nature 628, 450–457 (2024).
Kaake, R. M. et al. Characterization of an A3G-VifHIV-1-CRL5-CBFβ; structure using a cross-linking mass spectrometry pipeline for integrative modeling of host-pathogen complexes. Mol. Cell. Proteomics 20, 100132 (2021).
Kuenze, G. et al. Allosteric mechanism for KCNE1 modulation of KCNQ1 potassium channel activation. eLife 9, e57680 (2020).
Sun, J. & MacKinnon, R. Structural basis of human KCNQ1 modulation and gating. Cell 180, 340–347 e349 (2020).
Escobar, C. A. et al. Structural interactions define assembly adapter function of a type II secretion system pseudopilin. Structure 29, 1116–1127 e1118 (2021).
Banchenko, S. et al. Structural insights into Cullin4-RING ubiquitin ligase remodelling by Vpr from simian immunodeficiency viruses. PLoS Pathog. 17, e1009775 (2021).
Wheat, A. et al. Protein interaction landscapes revealed by advanced in vivo cross-linking-mass spectrometry. Proc. Natl Acad. Sci. USA 118, e2023360118 (2021).
Chen, Y. et al. Targeted cross-linker delivery for the in situ mapping of protein conformations and interactions in mitochondria. Nat. Commun. 14, 3882 (2023).
Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021).
Merkley, E. D. et al. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 23, 747–759 (2014).
Yoshida, S. et al. Molecular chaperone TRAP1 regulates a metabolic switch between mitochondrial respiration and aerobic glycolysis. Proc. Natl Acad. Sci. USA 110, E1604–E1612 (2013).
Thomas, P. D. The gene ontology and the meaning of biological function. Methods Mol. Biol. 1446, 15–24 (2017).
Bulbarelli, A., Valentini, A., DeSilvestris, M., Cappellini, M. D. & Borgese, N. An erythroid-specific transcript generates the soluble form of NADH-cytochrome b5 reductase in humans. Blood 92, 310–319 (1998).
Shor, B. & Schneidman-Duhovny, D. CombFold: predicting structures of large protein assemblies using a combinatorial assembly algorithm and AlphaFold2. Nat. Methods 21, 477–487 (2024).
Liu, S. et al. PSP: million-level protein sequence dataset for protein structure prediction. Preprint at https://arxiv.org/abs/2206.12240 (2022).
Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Kahraman, A., Malmström, L. & Aebersold, R. Xwalk: computing and visualizing distances in cross-linking experiments. Bioinformatics 27, 2163–2164 (2011).
Abhinandan, K. R. & Martin, A. C. Analysis and improvements to Kabat and structurally correct numbering of antibody variable domains. Mol. Immunol. 45, 3832–3839 (2008).
Gydo, C. Pv. Z. & Alexandre, M. J. J. B. Fast and sensitive rigid-body fitting into cryo-EM density maps with PowerFit. AIMS Biophys. 2, 73–87 (2015).
Eldar, Y., Lindenbaum, M., Porat, M. & Zeevi, Y. Y. The farthest point strategy for progressive image sampling. IEEE Trans. Image Process. 6, 1305–1315 (1997).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mitternacht, S. FreeSASA: an open source C library for solvent accessible surface area calculations. F1000Res. 5, 189 (2016).
Xie, Y. et al. Datasets and model weights for GRASP. OSF https://doi.org/10.17605/OSF.IO/6KJUQ (2025).
Xie, Y. et al. Source code for GRASP. Zenodo https://doi.org/10.5281/zenodo.15347070 (2025).
Acknowledgements
This work was supported by the National Science and Technology Major Project (grant no. 2022ZD0115001 to Y.Q.G. and S. Liu), the National Natural Science Foundation of China (grant nos. 92353304 and T2495221 to Y.Q.G.) and New Cornerstone Science Foundation (grant no. NCI202305 to Y.Q.G.). We thank K. Stahl from Technische Universität Berlin for recommending AlphaLink configuration and providing simulated CASP15 restraints for debugging. We thank F. N. Hitawala from Johns Hopkins University for sharing the PDB IDs and sequence data of their curated benchmarking dataset. We thank Y. Kaiguang from Dalian Institute of Chemical Physics to provide guidance for usage of the targeted XL-MS in the mitochondria. We also thank Z. Zhu for helpful discussions on binding energy calculation.
Author information
Authors and Affiliations
Contributions
Y.Q.G. and S. Liu designed and developed overall concepts in the paper and supervised the project. Y.X., C.Z. and X.D. developed the GRASP and Combfit methods. M.W. and Y.H. trained and evaluated MEGAFold-Multimer. Y.X., C.Z., S. Li, X.D., Y.L. and Z.C. performed data collection, evaluation and analysis. X.Y., C.Z., S. Li and X.D. wrote the initial draft of the paper. All authors contributed ideas to the work and assisted in editing and revision.
Corresponding authors
Ethics declarations
Competing interests
Changping Laboratory and Huawei Technologies Co., Ltd are in the process of applying for a patent covering the GRASP method, which lists authors including S. Liu, Y.X., C.Z. and Y.Q.G. as inventors. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Arunima Singh, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs 1–24, Tables 1–7, Methods 1–7 and References.
Supplementary Data
Detailed evaluation results and analytical data for each case across the six datasets.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xie, Y., Zhang, C., Li, S. et al. Integrating diverse experimental information to assist protein complex structure prediction by GRASP. Nat Methods 22, 2362–2374 (2025). https://doi.org/10.1038/s41592-025-02820-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02820-1