Abstract
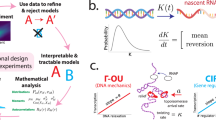
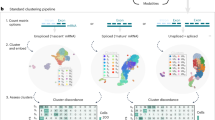
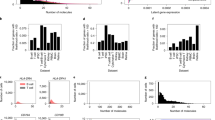
Single-cell RNA sequencing analysis centers on illuminating cell diversity and understanding the transcriptional mechanisms underlying cellular function. These datasets are large, noisy and complex. Current analyses prioritize noise removal and dimensionality reduction to tackle these challenges and extract biological insight. We propose an alternative, physical approach to leverage the stochasticity, size and multimodal nature of these data to explicitly distinguish their biological and technical facets while revealing the underlying regulatory processes. With the Python package Monod, we demonstrate how nascent and mature RNA counts, present in most published datasets, can be meaningfully ‘integrated’ under biophysical models of transcription. By using variation in these modalities, we can identify transcriptional modulation not discernible through changes in average gene expression, quantitatively compare mechanistic hypotheses of gene regulation, analyze transcriptional data from different technologies within a common framework and minimize the use of opaque or distortive normalization and transformation techniques.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets released by the Allen Institute for Brain Science were downloaded from http://data.nemoarchive.org/biccn/grant/u19_zeng/zeng/transcriptome/scell/10x_v3/mouse/raw/MOp/ and filtered according to the metadata annotations at http://data.nemoarchive.org/biccn/grant/u19_zeng/zeng/transcriptome/scell/10x_v3/mouse/processed/analysis/10X_cells_v3_AIBS/ refs. 56,136,155. The paired single-cell and single-nucleus mouse brain datasets as well as the human PBMC datasets were obtained from the 10x Genomics website. Control and IdU-perturbed mESC data47 were obtained from GSE176044 at the Gene Expression Omnibus. The mouse germ cell dataset57 was obtained from Gene Expression Omnibus repository GSE136220, the patient-derived PDAC tumor sample dataset61 was from GSE202051, and intestinal radiation therapy data60 were from GSE165318. Single-nucleus mESC data108 were obtained using Sequence Read Archive run accession number SRR18364193. Fluorescent intensity values for seqFISH+ experiments on mESCs107 were obtained from Zenodo (package 7693825)156. The prebuilt GRCh38 or mm10 genome from https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest (2020-A version) was used for quantification of datasets. All loom files with nascent and mature count matrices for the datasets used in this study and all Monod fits have been deposited in Zenodo (package 15051840)157,158.
Code availability
Notebooks that reproduce all the filtering, fitting and analysis procedures have been deposited also in Zenodo (package 15051840)157 and are available at https://github.com/pachterlab/monod_examples/tree/main/manuscript_computation. This GitHub repository also contains a Google Colaboratory notebook that illustrates the Monod workflow with a small dataset. The Monod software used for all analysis is available as a pip installable package with an API available at https://monod-examples.readthedocs.io.
References
Shekhar, K. et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323 (2016).
Yao, Z. et al. A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature 624, 317–332 (2023).
Wagner, D. E. et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018).
Regev, A. et al. Science forum: the human cell atlas. eLife 6, e27041 (2017).
Chari, T. et al. Whole-animal multiplexed single-cell RNA-seq reveals transcriptional shifts across Clytia medusa cell types. Sci. Adv. 7, eabh1683 (2021).
Luecken, M. D. & Theis, F. J. Current best practices in single cell RNA seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
Kharchenko, P. V. The triumphs and limitations of computational methods for scRNA-seq. Nat. Methods 18, 723–732 (2021).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 24, 550–572 (2023).
Kobak, D. & Berens, P. The art of using t-SNE for single-cell transcriptomics. Nat. Commun. 10, 5416 (2019).
Hu, Q. & Greene, C. S. Parameter tuning is a key part of dimensionality reduction via deep variational autoencoders for single cell RNA transcriptomics. Pac. Symp. Biocomput. 24, 362–373 (2019).
Raimundo, F., Vallot, C. & Vert, J.-P. Tuning parameters of dimensionality reduction methods for single-cell RNA-seq analysis. Genome Biol. 21, 212 (2020).
Cooley, S. M., Hamilton, T., Aragones, S. D., Ray, J. C. J. & Deeds, E. J. A novel metric reveals previously unrecognized distortion in dimensionality reduction of scRNA-seq data. Preprint at bioRxiv https://doi.org/10.1101/689851 (2020).
Chamberlin, J. T., Lee, Y., Marth, G. T. & Quinlan, A. R. Differences in molecular sampling and data processing explain variation among single-cell and single-nucleus RNA-seq experiments. Genome Res. 34, 179–188 (2024).
Ahlmann-Eltze, C. & Huber, W. Comparison of transformations for single-cell RNA-seq data. Nat. Methods 20, 665–672 (2023).
Chari, T. & Pachter, L. The specious art of single-cell genomics. PLoS Comput. Biol. 19, e1011288 (2023).
Gorin, G., Fang, M., Chari, T. & Pachter, L. RNA velocity unraveled. PLoS Comput. Biol. 18, e1010492 (2022).
Kiselev, V. Y., Andrews, T. S. & Hemberg, M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 20, 273–282 (2019).
Mimitou, E. P. et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol. 39, 1246–1258 (2021).
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Raser, J. M. & O’Shea, E. K. Noise in gene expression: origins, consequences, and control. Science 309, 2010–2013 (2005).
Gillespie, D. T. Stochastic simulation of chemical kinetics. Annu. Rev. Phys. Chem. 58, 35–55 (2007).
Guillemin, A. & Stumpf, M. P. H. Noise and the molecular processes underlying cell fate decision-making. Phys. Biol. 18, 011002 (2020).
Gorin, G., Vastola, J. J., Fang, M. & Pachter, L. Interpretable and tractable models of transcriptional noise for the rational design of single-molecule quantification experiments. Nat. Commun. 13, 7620 (2022).
Gorin, G. & Pachter, L. Length biases in single-cell RNA sequencing of pre-mRNA. Biophys. Rep. 3, 100097 (2023).
Gorin, G., Vastola, J. J. & Pachter, L. Studying stochastic systems biology of the cell with single-cell genomics data. Cell Syst. 14, 822–843 (2023).
Gorin, G. & Pachter, L. New and notable: revisiting the ‘two cultures’ through extrinsic noise. Biophys. J. 123, 1–3 (2024).
Raj, A., Peskin, C. S., Tranchina, D., Vargas, D. Y. & Tyagi, S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309 (2006).
Bokes, P., King, J. R., Wood, A. T. A. & Loose, M. Exact and approximate distributions of protein and mRNA levels in the low-copy regime of gene expression. J. Math. Biol. 64, 829–854 (2012).
Xu, H., Skinner, S. O., Sokac, A. M. & Golding, I. Stochastic kinetics of nascent RNA. Phys. Rev. Lett. 117, 128101 (2016).
Golding, I., Paulsson, J., Zawilski, S. M. & Cox, E. C. Real-time kinetics of gene activity in individual bacteria. Cell 123, 1025–1036 (2005).
Chari, T., Gorin, G. & Pachter, L. Biophysically interpretable inference of cell types from multimodal sequencing data. Nat. Comput. Sci. 4, 677–689 (2024).
Sullivan, D. K. et al. Accurate quantification of nascent and mature RNAs from single-cell and single-nucleus RNA-seq. Nucleic Acids Res. 53, gkae1137 (2024).
Sullivan, D. K. kallisto, bustools and kb-python for quantifying bulk, single-cell and single-nucleus RNA-seq. Nat. Protoc. 20, 587–607 (2025).
Jahnke, T. & Huisinga, W. Solving the chemical master equation for monomolecular reaction systems analytically. J. Math. Biol. 54, 1–26 (2006).
Vastola, J. J. Solving the chemical master equation for monomolecular reaction systems and beyond: a Doi–Peliti path integral view. J. Math. Biol. 83, 48 (2021).
Svensson, V. Droplet scRNA-seq is not zero-inflated. Nat. Biotechnol. 38, 147–150 (2020).
Singh, A. & Bokes, P. Consequences of mRNA transport on stochastic variability in protein levels. Biophys. J. 103, 1087–1096 (2012).
Dar, R. D. et al. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc. Natl Acad. Sci. USA 109, 17454–17459 (2012).
Halpern, K. B. et al. Bursty gene expression in the intact mammalian liver. Mol. Cell 58, 147–156 (2015).
Corrigan, A. M., Tunnacliffe, E., Cannon, D. & Chubb, J. R. A continuum model of transcriptional bursting. eLife 5, e13051 (2016).
Fukaya, T., Lim, B. & Levine, M. Enhancer control of transcriptional bursting. Cell 166, 358–368 (2016).
Nicolas, D., Phillips, N. E. & Naef, F. What shapes eukaryotic transcriptional bursting? Mol. Biosyst. 13, 1280–1290 (2017).
Rodriguez, J. & Larson, D. R. Transcription in living cells: molecular mechanisms of bursting. Annu. Rev. Biochem. 89, 189–212 (2020).
Ham, L., Brackston, R. D. & Stumpf, M. P. H. Extrinsic noise and heavy-tailed laws in gene expression. Phys. Rev. Lett. 124, 108101 (2020).
Gorin, G., Yoshida, S. & Pachter, L. Assessing Markovian and delay models for single-nucleus RNA sequencing. Bull. Math. Biol. 85, 114 (2023).
Leier, A. & Marquez-Lago, T. T. Delay chemical master equation: direct and closed-form solutions. Proc. R. Soc. Math. Phys. Eng. Sci. 471, 20150049 (2015).
Desai, R. V. et al. A DNA repair pathway can regulate transcriptional noise to promote cell fate transitions. Science 373, eabc6506 (2021).
Hilfinger, A. & Paulsson, J. Separating intrinsic from extrinsic fluctuations in dynamic biological systems. Proc. Natl Acad. Sci. USA 108, 12167–12172 (2011).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Hansen, M. M. K., Desai, R. V., Simpson, M. L. & Weinberger, L. S. Cytoplasmic amplification of transcriptional noise generates substantial cell-to-cell variability. Cell Syst. 7, 384–397 (2018).
García-Blay, Ó. et al. Multimodal screen identifies noise-regulatory proteins. Dev. Cell 60, 133–151 (2025).
Calia, G. P., Chen, X., Zuckerman, B. & Weinberger, L. S. Comparative analysis between single-cell RNA-seq and single-molecule RNA FISH indicates that the pyrimidine nucleobase idoxuridine (IdU) globally amplifies transcriptional noise. Preprint at bioRxiv https://doi.org/10.1101/2023.03.14.532632 (2023).
Huang, Z. et al. Deep learning linking mechanistic models to single-cell transcriptomics data reveals transcriptional bursting in response to DNA damage. eLife https://doi.org/10.7554/eLife.100623.2 (2024).
Yao, Z. et al. A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature 598, 103–110 (2021).
Mayère, C. et al. Single-cell transcriptomics reveal temporal dynamics of critical regulators of germ cell fate during mouse sex determination. FASEB J. 35, e21452 (2021).
Stelzer, G. et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics 54, 1.30.1–1.30.33 (2016).
Lee, S. C.-W. & Abdel-Wahab, O. Therapeutic targeting of splicing in cancer. Nat. Med. 22, 976–986 (2016).
Lu, H. et al. Single-cell map of dynamic cellular microenvironment of radiation-induced intestinal injury. Commun. Biol. 6, 1248 (2023).
Hwang, W. L. et al. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nat. Genet. 54, 1178–1191 (2022).
Dyson, N. J. RB1: a prototype tumor suppressor and an enigma. Genes Dev. 30, 1492–1502 (2016).
Touil, Y. et al. Colon cancer cells escape 5FU chemotherapy-induced cell death by entering stemness and quiescence associated with the c-Yes/YAP axis. Clin. Cancer Res. 20, 837–846 (2014).
Huang, X. et al. Identification of genes related to 5-fluorouracil based chemotherapy for colorectal cancer. Front. Immunol. 13, 887048 (2022).
Sahebnasagh, R., Azizi, Z., Komeili-Movahhed, T., Zendehdel, K. & Ghahremani, M. H. In-silico and in-vitro investigation of key long non-coding RNAs involved in 5-fluorouracil resistance in colorectal cancer cells: analyses highlighting NEAT1 and MALAT1 as contributors. Cureus 16, e66393 (2024).
Machkalyan, G., Hèbert, T. E. & Miller, G. J. PPIP5K1 suppresses etoposide-triggered apoptosis. J. Mol. Signal. 11, 4 (2016).
Bhandari, K. & Ding, W.-Q. Protein arginine methyltransferases in pancreatic ductal adenocarcinoma: new molecular targets for therapy. Int. J. Mol. Sci. 25, 3958 (2024).
Wang, X. et al. Characterization of LIMA1 and its emerging roles and potential therapeutic prospects in cancers. Front. Oncol. 13, 1115943 (2023).
Hashemzehi, M. et al. Angiotensin receptor blocker losartan inhibits tumor growth of colorectal cancer. EXCLI J. 20, 506–521 (2021).
Pothula, S. P. et al. Targeting HGF/c-MET axis in pancreatic cancer. Int. J. Mol. Sci. 21, 9170 (2020).
Leclerc, G. J., Leclerc, G. M. & Barredo, J. C. Real-time RT–PCR analysis of mRNA decay: half-life of β-actin mRNA in human leukemia CCRF-CEM and Nalm-6 cell lines. Cancer Cell Int. 2, 1 (2002).
Izdebska, M., Zielińska, W., Hałas-Wiśniewska, M. & Grzanka, A. Involvement of actin and actin-binding proteins in carcinogenesis. Cells 9, 2245 (2020).
Arina, A. et al. Tumor-reprogrammed resident T cells resist radiation to control tumors. Nat. Commun. 10, 3959 (2019).
Shadad, A. K., Sullivan, F. J., Martin, J. D. & Egan, L. J. Gastrointestinal radiation injury: symptoms, risk factors and mechanisms. World J. Gastroenterol. 19, 185–198 (2013).
Augustin, R. C., Bao, R. & Luke, J. J. Targeting Cbl-b in cancer immunotherapy. J. Immunother. Cancer 11, e006007 (2023).
Yu, M. et al. CD73 on cancer-associated fibroblasts enhanced by the A2B-mediated feedforward circuit enforces an immune checkpoint. Nat. Commun. 11, 515 (2020).
Qu, L. et al. NCAPD3 is a prognostic biomarker and is correlated with immune infiltrates in glioma. Histol. Histopathol. 39, 1473–1484 (2024).
Zhong, Y. et al. Insulin-like growth factor 2 receptor is a key immune-related gene that is correlated with a poor prognosis in patients with triple-negative breast cancer: a bioinformatics analysis. Front. Oncol. 12, 871786 (2022).
Xu, X. et al. Pan-cancer analysis of the role of MPP7 in human tumors. Heliyon 10, e36148 (2024).
Dittmer, J. The biology of the Ets1 proto-oncogene. Mol. Cancer 2, 29 (2003).
Phee, H., Mollenauer, M. N. & Weiss, A. Role of GIT2 in T cell migration and development (95.8). J. Immunol. 182, 95.8 (2009).
O’Hagan, K. L., Miller, S. & Phee, H. Pak2 is essential for the function of Foxp3+ regulatory T cells through maintaining a suppressive Treg phenotype. Sci. Rep. 7, 17097 (2017).
Belenguer, G. et al. RNF43/ZNRF3 loss predisposes to hepatocellular-carcinoma by impairing liver regeneration and altering the liver lipid metabolic ground-state. Nat. Commun. 13, 334 (2022).
Yue, F. et al. Loss of ZNRF3/RNF43 unleashes EGFR in cancer. eLife https://doi.org/10.7554/eLife.95639.2 (2024).
Wei, J.-L. et al. GCH1 induces immunosuppression through metabolic reprogramming and IDO1 upregulation in triple-negative breast cancer. J. Immunother. Cancer 9, e002383 (2021).
Zeng, Q. et al. LCP1 is a prognostic biomarker correlated with immune infiltrates in gastric cancer. Cancer Biomark. 30, 105–125 (2021).
Pan, S. et al. Decreased expression of ARHGAP15 promotes the development of colorectal cancer through PTEN/AKT/FOXO1 axis. Cell Death Dis. 9, 673 (2018).
Dou, B., Jiang, G., Peng, W. & Liu, C. OTULIN deficiency: focus on innate immune system impairment. Front. Immunol. 15, 1371564 (2024).
Wang, W.-Y. et al. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 16, 1383–1391 (2013).
Martínez, D. et al. Discovery of BbX transcription factor in the Patagonian blennie: exploring expression changes following combined bacterial and thermal stress exposure. Dev. Comp. Immunol. 149, 105056 (2023).
Kim, T. H., Zhou, X. & Chen, M. Demystifying ‘drop-outs’ in single-cell UMI data. Genome Biol. 21, 196 (2020).
Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746 (2020).
Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017).
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019).
Bennett, H. M., Stephenson, W., Rose, C. M. & Darmanis, S. Single-cell proteomics enabled by next-generation sequencing or mass spectrometry. Nat. Methods 20, 363–374 (2023).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Minnoye, L. et al. Chromatin accessibility profiling methods. Nat. Rev. Methods Primers 1, 10 (2021).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Ashuach, T., Reidenbach, D. A., Gayoso, A. & Yosef, N. PeakVI: a deep generative model for single-cell chromatin accessibility analysis. Cell Rep. Methods 2, 100182 (2022).
Ashuach, T. et al. MultiVI: deep generative model for the integration of multimodal data. Nat. Methods 20, 1222–1231 (2023).
Tyler, S. R., Guccione, E. & Schadt, E. E. Erasure of biologically meaningful signal by unsupervised scRNAseq batch-correction methods. Preprint at bioRxiv https://doi.org/10.1101/2021.11.15.468733 (2023).
Shah, S. et al. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174, 363–376 (2018).
Takei, Y. et al. High-resolution spatial multi-omics reveals cell-type specific nuclear compartments. Preprint at bioRxiv https://doi.org/10.1101/2023.05.07.539762 (2023).
Khateb, M. et al. Transcriptomics, regulatory syntax, and enhancer identification in mesoderm-induced ESCs at single-cell resolution. Cell Rep. 40, 111219 (2022).
10x Genomics. 30k Mouse E18 Combined Cortex, Hippocampus and Subventricular Zone Cells Multiplexed, 12 CMOs, Brain 4 https://www.10xgenomics.com/datasets/30-k-mouse-e-18-combined-cortex-hippocampus-and-subventricular-zone-cells-multiplexed-12-cm-os-3-1-standard-6-0-0 (2021).
10x Genomics. 30k Mouse E18 Combined Cortex, Hippocampus and Subventricular Zone Nuclei Multiplexed, 12 CMOs, Brain Nuclei 4 https://www.10xgenomics.com/datasets/30-k-mouse-e-18-combined-cortex-hippocampus-and-subventricular-zone-nuclei-multiplexed-12-cm-os-3-1-standard-6-0-0 (2021).
Swain, P. S., Elowitz, M. B. & Siggia, E. D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl Acad. Sci. USA 99, 12795–12800 (2002).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at arxiv.org/abs/1802.03426 (2018).
Grima, R. & Esmenjaud, P.-M. Quantifying and correcting bias in transcriptional parameter inference from single-cell data. Biophys. J. 123, 4–30 (2024).
Cao, Z. & Grima, R. Linear mapping approximation of gene regulatory networks with stochastic dynamics. Nat. Commun. 9, 3305 (2018).
Gorin, G. & Pachter, L. Modeling bursty transcription and splicing with the chemical master equation. Biophys. J. 121, 1056–1069 (2022).
Felce, C., Gorin, G. & Pachter, L. Biophysical model for joint analysis of chromatin and RNA sequencing data. Phys. Rev. E 110, 064405 (2024).
Alpert, T., Herzel, L. & Neugebauer, K. M. Perfect timing: splicing and transcription rates in living cells. Wiley Interdiscip. Rev. RNA 8, e1401 (2017).
Herzog, V. A. et al. Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 14, 1198–1204 (2017).
Gorin, G., Carilli, M., Chari, T. & Pachter, L. Spectral neural approximations for models of transcriptional dynamics. Biophys. J. 123, 2892–2901 (2024).
Sukys, A., Öcal, K. & Grima, R. Approximating solutions of the chemical master equation using neural networks. iScience 25, 105010 (2022).
Cao, Z. et al. Efficient and scalable prediction of stochastic reaction–diffusion processes using graph neural networks. Math. Biosci. 375, 109248 (2024).
Carilli, M. T., Gorin, G., Choi, Y., Chari, T. & Pachter, L. Biophysical modeling with variational autoencoders for bimodal, single-cell RNA sequencing data. Nat. Methods 21, 1466–1469 (2024).
Fang, M., Gorin, G. & Pachter, L. Trajectory inference from single-cell genomics data with a process time model. PLoS Comp. Biol. 21, e1012752 (2024).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Brodtkorb, P. A. & D’Errico, J. numdifftools https://pypi.org/project/numdifftools/ (2021).
Melsted, P. et al. Modular, efficient and constant-memory single-cell RNA-seq preprocessing. Nat. Biotechnol. 39, 813–818 (2021).
Gorin, F. & Pachter, L. Intrinsic and extrinsic noise are distinguishable in a synthesis – export – degradation model of mRNA production. Preprint at bioRxiv https://doi.org/10.1101/2020.09.25.312868 (2020).
Ham, L., Schnoerr, D., Brackston, R. D. & Stumpf, M. P. H. Exactly solvable models of stochastic gene expression. J. Chem. Phys. 152, 144106 (2020).
Ham, L., Jackson, M. & Stumpf, M. P. H. Pathway dynamics can delineate the sources of transcriptional noise in gene expression. eLife 10, e69324 (2021).
Jiang, Q. et al. Neural network aided approximation and parameter inference of non-Markovian models of gene expression. Nat. Commun. 12, 2618 (2021).
The MathWorks. MATLAB R2022a Symbolic Math Toolbox (MathWorks, 2022).
The MathWorks. MATLAB R2022a (MathWorks, 2022).
Tang, W., Jørgensen, A. C. S., Marguerat, S., Thomas, P. & Shahrezaei, V. Modelling capture efficiency of single cell RNA-sequencing data improves inference of transcriptome-wide burst kinetics. Bioinformatics 39, btad395 (2023).
Munsky, B., Li, G., Fox, Z. R., Shepherd, D. P. & Neuert, G. Distribution shapes govern the discovery of predictive models for gene regulation. Proc. Natl Acad. Sci. USA 115, 7533–7538 (2018).
Allen Institute for Brain Science. FASTQ Files for Allen v3 Mouse MOp Samples http://data.nemoarchive.org/biccn/grant/u19_zeng/zeng/transcriptome/scell/10x_v3/mouse/raw/MOp (2020).
Melsted, P. áll., Ntranos, V. & Pachter, L. The barcode, UMI, set format and BUStools. Bioinformatics 35, 4472–4473 (2019).
Burnham, K. P. & Anderson, D. R. Model Selection and Multimodel Inference: a Practical Information–Theoretic Approach 2nd edn (Springer, 2002).
Ly, A., Marsman, M., Verhagen, J., Grasman, R. P. P. P. & Wagenmakers, E.-J. A tutorial on Fisher information. J. Math. Psychol. 80, 40–55 (2017).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Halpern, K. B. et al. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 13, 2653–2662 (2015).
McKellar, D. W. et al. Large-scale integration of single-cell transcriptomic data captures transitional progenitor states in mouse skeletal muscle regeneration. Commun. Biol. 4, 1280 (2021).
Jew, B. et al. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 11, 1971 (2020).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Montserrat Ayuso, T. & Esteve-Codina, A. Revealing the prevalence of suboptimal cells and organs in reference cell atlases: an imperative for enhanced quality control. BMC Genomics 25, 1124 (2024).
Clarke, Z. A. & Bader, G. MALAT1 expression indicates cell quality in single-cell RNA sequencing data. Preprint at bioRxiv https://doi.org/10.1101/2024.07.14.603469 (2024).
Pratapa, A., Jalihal, A. P., Law, J. N., Bharadwaj, A. & Murali, T. M. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods 17, 147–154 (2020).
Kim, D. et al. Gene regulatory network reconstruction: harnessing the power of single-cell multi-omic data. NPJ Syst. Biol. Appl. 9, 51 (2023).
Stumpf, M. P. H. Statistical and computational challenges for whole cell modelling. Curr. Opin. Syst. Biol. 26, 58–63 (2021).
Saint-Antoine, M. M. & Singh, A. Network inference in systems biology: recent developments, challenges, and applications. Curr. Opin. Biotechnol. 63, 89–98 (2020).
Stumpf, M. P. H. Inferring better gene regulation networks from single-cell data. Curr. Opin. Syst. Biol. 27, 100342 (2021).
10x Genomics. 1k PBMCs from a Healthy Donor (v2 Chemistry), Single Cell Gene Expression Dataset by Cell Ranger 3.0.0 https://www.10xgenomics.com/datasets/1-k-pbm-cs-from-a-healthy-donor-v-2-chemistry-3-standard-3-0-0 (2018).
10x Genomics. 1k PBMCs from a Healthy Donor (v3 Chemistry), Single Cell Gene Expression Dataset by Cell Ranger 3.0.0 https://www.10xgenomics.com/datasets/1-k-pbm-cs-from-a-healthy-donor-v-3-chemistry-3-standard-3-0-0 (2018).
Allen Institute for Brain Science. Analyses for Allen v3 Mouse MOp Samples http://data.nemoarchive.org/biccn/grant/u19_zeng/zeng/transcriptome/scell/10x_v3/mouse/processed/analysis/10X_cells_v3_AIBS/ (2020).
Takei, Y., Yang, Y. & Cai, L. High-resolution spatial multi-omics datasets. Zenodo https://doi.org/10.5281/zenodo.7693825 (2023).
Gorin, G., Pachter, L., Chari, T., Carilli, M. & Vastola, J. Monod supporting data. Zenodo https://doi.org/10.5281/zenodo.15051840 (2025).
Jayakumar, K. & Mundassery, D. A. On Moran’s bivariate gamma and bivariate negative binomial distribution. Calcutta Stat. Assoc. Bull. 59, 15–28 (2007).
Acknowledgements
We thank S. Booeshaghi and M. Fang for useful discussions in the course of developing Monod. G.G. and L.P. were partially funded by NIH U19MH114830 and NIH 5UM1HG012077-02. M.C. is supported by the National Science Foundation Graduate Research Fellowship Program under grant no. 2139433. T.C. was supported by the Eric and Wendy Schmidt Center at the Broad Institute of MIT and Harvard. Part of this work was performed during G.G.’s Data Sciences Co-op with Celsius Therapeutics.
Author information
Authors and Affiliations
Contributions
All authors contributed extensively to the work presented in this paper. The first draft of the paper was conceptualized and written by G.G. and L.P. ‘Assumptions and approach of Monod’ was predominantly conceptualized by G.G., J.J.V. and L.P. ‘Monod generalizes differential expression analyses’ was predominantly conceptualized and executed by G.G., T.C. and L.P. ‘Monod identifies strategies of resistance and recovery’ and ‘Model selection and insight about gene regulation strategies’ were predominantly conceptualized and executed by M.C. and L.P. ‘Principled integration of multiple modalities’ was predominantly conceptualized and executed by G.G., J.J.V., M.C. and L.P. ‘Assessing loss of biological signal after preprocessing’ was predominantly conceptualized and executed by G.G., J.J.V. and L.P.
Corresponding author
Ethics declarations
Competing interests
G.G. is an employee of Fauna Bio. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Lin Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Tables 1–10 and Figs. 1–19
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gorin, G., Chari, T., Carilli, M. et al. Monod: model-based discovery and integration through fitting stochastic transcriptional dynamics to single-cell sequencing data. Nat Methods 22, 2286–2300 (2025). https://doi.org/10.1038/s41592-025-02832-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02832-x