Abstract
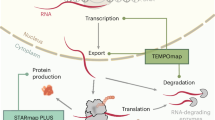
Characterizing the transcriptional and translational gene expression patterns at the single-cell level within their three-dimensional (3D) tissue context is essential for revealing how genes shape tissue structure and function in health and disease. However, most existing spatial profiling techniques are limited to 5–20 µm thin tissue sections. Here, we developed Deep-STARmap and Deep-RIBOmap, which enable 3D in situ quantification of thousands of gene transcripts and their corresponding translation activities, respectively, within 60–200-µm thick tissue blocks. This is achieved through scalable probe synthesis, hydrogel embedding with efficient probe anchoring and robust cDNA crosslinking. We first utilized Deep-STARmap in combination with multicolor fluorescent protein imaging for simultaneous molecular cell typing and 3D neuron morphology tracing in the mouse brain. We also demonstrate that 3D spatial profiling facilitates comprehensive and quantitative analysis of tumor–immune interactions in human skin cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Deep-STARmap and Deep-RIBOmap sequencing datasets of 1,017-gene in mouse brain, Deep-STARmap datasets of 254-gene in human cutaneous squamous cell carcinoma tissue are available in Zenodo at https://doi.org/10.5281/zenodo.16783354 (ref. 85). The mouse brain atlas dataset used is available at https://singlecell.broadinstitute.org/single_cell/study/SCP1830. The cSCC scRNA-seq dataset used is available at the Gene Expression Omnibus under accession code GSE144240.
Code availability
Deep-STARmap and Deep-RIBOmap are implemented based on MATLAB v.R2023b, Python v.3.8 and R v.4.2.1. The data processing tool has been deposited in Zenodo at https://doi.org/10.5281/zenodo.16783354 (ref. 85). Additional information is available at the Wang laboratory website at https://www.wangxiaolab.org/.
References
Tabula Sapiens Consortium et al. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022).
Elmentaite, R., Domínguez Conde, C., Yang, L. & Teichmann, S. A. Single-cell atlases: shared and tissue-specific cell types across human organs. Nat. Rev. Genet. 23, 395–410 (2022).
Hwang, B., Lee, J. H. & Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 50, 1–14 (2018).
Baysoy, A., Bai, Z., Satija, R. & Fan, R. The technological landscape and applications of single-cell multi-omics. Nat. Rev. Mol. Cell Biol. 24, 695–713 (2023).
Brar, G. A. & Weissman, J. S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 16, 651–664 (2015).
Ingolia, N. T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 15, 205–213 (2014).
Vandereyken, K., Sifrim, A., Thienpont, B. & Voet, T. Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 24, 494–515 (2023).
Zhuang, X. Spatially resolved single-cell genomics and transcriptomics by imaging. Nat. Methods 18, 18–22 (2021).
Lein, E., Borm, L. E. & Linnarsson, S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358, 64–69 (2017).
Liao, J., Lu, X., Shao, X., Zhu, L. & Fan, X. Uncovering an organ’s molecular architecture at single-cell resolution by spatially resolved transcriptomics. Trends Biotechnol. 39, 43–58 (2021).
Ren, J., Luo, S., Shi, H. & Wang, X. Spatial omics advances for in situ RNA biology. Mol. Cell 84, 3737–3757 (2024).
Richardson, D. S. & Lichtman, J. W. Clarifying tissue clearing. Cell 162, 246–257 (2015).
Ueda, H. R. et al. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 21, 61–79 (2020).
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
Sakaguchi, R., Leiwe, M. N. & Imai, T. Bright multicolor labeling of neuronal circuits with fluorescent proteins and chemical tags. eLife https://doi.org/10.7554/elife.40350 (2018).
Kebschull, J. M. et al. High-throughput mapping of single-neuron projections by sequencing of barcoded RNA. Neuron 91, 975–987 (2016).
Chen, X. et al. High-throughput mapping of long-range neuronal projection using in situ sequencing. Cell 179, 772–786.e19 (2019).
Jun, J. J. et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236 (2017).
Manz, K. M., Siemann, J. K., McMahon, D. G. & Grueter, B. A. Patch-clamp and multi-electrode array electrophysiological analysis in acute mouse brain slices. STAR Protoc. 2, 100442 (2021).
Li, Q. et al. Multimodal charting of molecular and functional cell states via in situ electro-sequencing. Cell 186, 2002–2017.e21 (2023).
Gouwens, N. W. et al. Integrated morphoelectric and transcriptomic classification of cortical GABAergic cells. Cell 183, 935–953.e19 (2020).
Fuzik, J. et al. Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat. Biotechnol. 34, 175–183 (2016).
Cadwell, C. R. et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 34, 199–203 (2016).
Xu, S. et al. Behavioral state coding by molecularly defined paraventricular hypothalamic cell type ensembles. Science https://doi.org/10.1126/science.abb2494 (2020).
Yang, W. & Yuste, R. In vivo imaging of neural activity. Nat. Methods 14, 349–359 (2017).
Bugeon, S. et al. Publisher correction: a transcriptomic axis predicts state modulation of cortical interneurons. Nature 609, E10 (2022).
Almagro, J., Messal, H. A., Zaw Thin, M., van Rheenen, J. & Behrens, A. Tissue clearing to examine tumour complexity in three dimensions. Nat. Rev. Cancer 21, 718–730 (2021).
Fang, R. et al. Three-dimensional single-cell transcriptome imaging of thick tissues. eLife https://doi.org/10.7554/eLife.90029 (2023).
Wang, Y. et al. EASI-FISH for thick tissue defines lateral hypothalamus spatio-molecular organization. Cell 184, 6361–6377.e24 (2021).
Gandin, V. et al. Deep-tissue transcriptomics and subcellular imaging at high spatial resolution. Science 388, eadq2084 (2025).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science https://doi.org/10.1126/science.aat5691 (2018).
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Kanatani, S. et al. Whole-brain spatial transcriptional analysis at cellular resolution. Science 386, 907–915 (2024).
Clarke, D. N., Formery, L. & Lowe, C. J. See-Star: a versatile hydrogel-based protocol for clearing large, opaque and calcified marine invertebrates. Evodevo 15, 8 (2024).
Choi, H. M. T. et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 28, 1208–1212 (2010).
Choi, H. M. T., Beck, V. A. & Pierce, N. A. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano. https://doi.org/10.1021/nn405717p (2014).
Shah, S. et al. Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862–2867 (2016).
Choi, H. M. T. et al. Mapping a multiplexed zoo of mRNA expression. Development 143, 3632–3637 (2016).
Choi, H. M. T. et al. Third-generation hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development https://doi.org/10.1242/dev.165753 (2018).
Yang, B. et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014).
Sylwestrak, E. L., Rajasethupathy, P., Wright, M. A., Jaffe, A. & Deisseroth, K. Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell 164, 792–804 (2016).
Yoshimura, Y. & Fujimoto, K. Ultrafast reversible photo-cross-linking reaction: toward in situ DNA manipulation. Org. Lett. 10, 3227–3230 (2008).
Yoshimura, Y., Ohtake, T., Okada, H. & Fujimoto, K. A new approach for reversible RNA photocrosslinking reaction: application to sequence-specific RNA selection. ChemBioChem 10, 1473–1476 (2009).
Fujimoto, K., Konishi-Hiratsuka, K., Sakamoto, T. & Yoshimura, Y. Site-specific cytosine to uracil transition by using reversible DNA photo-crosslinking. ChemBioChem 11, 1661–1664 (2010).
Chen, F. et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684 (2016).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Zeng, H. et al. Spatially resolved single-cell translatomics at molecular resolution. Science 380, eadd3067 (2023).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018).
Saunders, A. et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030.e16 (2018).
Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018).
Shi, H. et al. Spatial atlas of the mouse central nervous system at molecular resolution. Nature 622, 552–561 (2023).
He, Y. et al. Towards a universal spatial molecular atlas of the mouse brain. Preprint at bioRxiv https://doi.org/10.1101/2024.05.27.594872 (2024).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Dong, H. W. & The Allen Institute for Brain Science. The Allen Reference Atlas, (Book + CD-ROM): A Digital Color Brain Atlas of the C57BL/6J Male Mouse (Wiley, 2008).
Legland, D., Arganda-Carreras, I. & Andrey, P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32, 3532–3534 (2016).
Amitai, Y. et al. The spatial dimensions of electrically coupled networks of interneurons in the neocortex. J. Neurosci. 22, 4142–4152 (2002).
Ebina, T. et al. 3D clustering of GABAergic neurons enhances inhibitory actions on excitatory neurons in the mouse visual cortex. Cell Rep. 9, 1896–1907 (2014).
Gibson, J. R., Beierlein, M. & Connors, B. W. Two networks of electrically coupled inhibitory neurons in neocortex. Nature 402, 75–79 (1999).
Xu, C. S. et al. Enhanced FIB-SEM systems for large-volume 3D imaging. eLife https://doi.org/10.7554/elife.25916 (2017).
Scheffer, L. K. et al. A connectome and analysis of the adult central brain. eLife https://doi.org/10.7554/elife.57443 (2020).
Cai, D., Cohen, K. B., Luo, T., Lichtman, J. W. & Sanes, J. R. Improved tools for the Brainbow toolbox. Nat. Methods 10, 540–547 (2013).
Chan, K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017).
Spruston, N. Pyramidal neurons: dendritic structure and synaptic integration. Nat. Rev. Neurosci. 9, 206–221 (2008).
Jiang, S. et al. Anatomically revealed morphological patterns of pyramidal neurons in layer 5 of the motor cortex. Sci. Rep. 10, 7916 (2020).
Zeng, H. What is a cell type and how to define it?. Cell 185, 2739–2755 (2022).
Guzman, A. K., Schmults, C. D. & Ruiz, E. S. Squamous cell carcinoma: an update in staging, management, and postoperative surveillance strategies. Dermatol. Clin. 41, 1–11 (2023).
Borden, E. S. et al. Neoantigen fitness model predicts lower immune recognition of cutaneous squamous cell carcinomas than actinic keratoses. Front. Immunol. 10, 2799 (2019).
Levine, D. E., Karia, P. S. & Schmults, C. D. Outcomes of patients with multiple cutaneous squamous cell carcinomas: a 10-year single-institution cohort study. JAMA Dermatol. 151, 1220–1225 (2015).
Gonzalez, J. L. et al. Multiple cutaneous squamous cell carcinoma in immunosuppressed vs immunocompetent patients. JAMA Dermatol. 155, 625–627 (2019).
Migden, M. R. et al. PD-1 blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N. Engl. J. Med. 379, 341–351 (2018).
Joost, S. et al. The molecular anatomy of mouse skin during hair growth and rest. Cell Stem Cell 26, 441–457.e7 (2020).
Ji, A. L. et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182, 1661–1662 (2020).
Jerby-Arnon, L. et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 175, 984–997.e24 (2018).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
He, Y. et al. ClusterMap for multi-scale clustering analysis of spatial gene expression. Nat. Commun. 12, 5909 (2021).
Fujita, H. et al. Langerhans cells from human cutaneous squamous cell carcinoma induce strong type 1 immunity. J. Invest. Dermatol. 132, 1645–1655 (2012).
Klechevsky, E. et al. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity 29, 497–510 (2008).
Calà, G., Sina, B., De Coppi, P., Giobbe, G. G. & Gerli, M. F. M. Primary human organoids models: current progress and key milestones. Front. Bioeng. Biotechnol. 11, 1058970 (2023).
Ren, J. et al. Spatiotemporally resolved transcriptomics reveals the subcellular RNA kinetic landscape. Nat. Methods 20, 695–705 (2023).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Palla, G. et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods 19, 171–178 (2022).
Sui, X. et al. Scalable spatial single-cell transcriptomics and translatomics in 3D thick tissue blocks. Zenodo https://doi.org/10.5281/zenodo.16783354 (2025).
Acknowledgements
The authors thank the B. E. Deverman laboratory (Stanley Center for Psychiatric Research, Broad Institute) for helping with AAV packages and V. Neel (Department of Dermatology, Massachusetts General Hospital) for providing patient SCC samples critical to this study. The authors thank H. Shi, A. Roy, F. Kostas and S. Furniss (Broad Institute) for their helpful feedback and comments during manuscript writing. We thank P. Paroutis and the Hospital for Sick Children imaging facility for software access. X.W. gratefully acknowledges support from Edward Scolnick Professorship, gifts from Stanley Center for Psychiatric Research, Escaping Velocity Award, Ono Pharma Breakthrough Science Initiative Award, Merkin Institute Fellowship and National Institutes of Health (NIH) DP2 New Innovator Award (1DP2GM146245) and NIH BRAIN CONNECTS (UM1 NS132173). J.A.L. gratefully acknowledges support from the NIH (T32AR007098-47), Dermatology Foundation (Dermatologist Investigator Research Fellowship and Physician-Scientist Career Development Award), and BroadIgnite. W.X.W. is a Damon Runyon–National Mah Jongg League Fellow, supported by the Damon Runyon Cancer Research Foundation (DRG no. 2512-23). D.L.B. is supported by funding from the Eric and Wendy Schmidt Center at the Broad Institute of MIT and Harvard. J.L. acknowledges the support of NIH/NIDDK 1DP1DK130673. Figures include graphics created in BioRender (https://BioRender.com/ew2lkjv).
Author information
Authors and Affiliations
Contributions
X.S. and X.W. conceived the idea and developed Deep-STARmap and Deep-RIBOmap for the study. X.S. carried out experimental work, performed in situ sequencing and conducted computational and data analyses. J.A.L. designed the gene lists for human cSCC, acquired samples and made significant contributions to the analysis of human cSCC. S.L., Y.H. and Z.T. performed data analysis. Z.L. helped with method optimization. Y.Z. helped with AAV packaging and conducted animal work. W.X.W. helped with morphology analysis. D.L.B. conducted prediction of molecular cell type based on neuronal morphology. J.L. provided critical discussions during method development. X.S., J.A.L. and X.W. wrote the manuscript with input from all authors. X.W. supervised the study.
Corresponding author
Ethics declarations
Competing interests
X.W. and X.S. are inventors on pending patent applications related to Deep-STARmap and Deep-RIBOmap. X.W. is a scientific co-founder and consultant of Stellaromics. All other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Lei Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Optimization of probe crosslinking.
a, Representative fluorescent imaging illustrating probe anchoring efficiency in a hydrogel matrix with various adaptor-primer ratios. Red: DNA amplicons of brain cell type markers: Gfap, Snap25, C1qa, Plp1. Blue: DAPI. Scale bar: 10 µm. n = 4 technical replicates, each derived from independent cortical regions across four different brain slices. b, 15% TBE-Urea gels demonstrating UV crosslinking efficiency with varying adaptor-primer molar ratios. CNVK- and Acrydite-containing adaptor used for UV crosslinking is [5Acryd]GCTA[cnvK]ATACGTCGTACTGGTAGG[Inv-dT] (24 nt). Primer used is 58 bp ssDNA with a 24 nt flanking liner at the 5′ end. M, Marker: IDT ssDNA 20/100 Ladder. n = 3 replicates. c, Quantification of cell images showing the average amplicon reads per cell (n = 4 technical replicates, each derived from independent cortical regions across four different brain slices). Two-sided independent t-test, ***P < 0.001, ****P < 0.0001. Data presented as mean ± standard deviation. d, Representative fluorescent imaging demonstrating detection efficiency of covalently anchored RNA molecules or probes within the hydrogel in the Deep-STARmap setting. Red: DNA amplicons from 4 cell type markers (Supplementary Table 1). Blue: DAPI. Scale bar: 50 µm. e, Quantification of cell images showing the average amplicon reads per cell (n = 4 technical replicates, each derived from independent cortical regions across four different brain slices). Two-sided independent t-test, ****P < 0.0001. Data presented as mean ± standard deviation. f, Representative fluorescent images of ACTB-gene profiling in tissue sections using Deep-STARmap with an Acrydite-modified adaptor (left) and an Acrydite-modified poly-dT oligo (right). Red: DNA amplicons, blue: DAPI nuclear staining. Scale bar: 40 µm. g, Box plot showing the number of reads per cell when using an Acrydite-modified adaptor (top) versus an Acrydite-modified poly-dT oligo (bottom) for anchoring in the Deep-STARmap workflow. n = 4 technical replicates, each derived from independent cortical regions across four different brain slices. Two-sided independent t-test, ***P < 0.001. Data are shown as mean ± standard deviation. h, Representative fluorescent imaging demonstrating probe anchoring efficiency with (left) and without (right) the use of the VA-044 thermal initiator in the first round of polymerization. Red: DNA amplicons from 4 cell type markers (Supplementary Table 1). Blue: DAPI. Scale bar: 50 µm. i, Quantification of cell images showing the average amplicon reads per cell (n = 4 technical replicates, each derived from independent cortical regions across four different brain slices). Two-sided independent t-test, ****P < 0.0001. Data presented as mean ± standard deviation.
Extended Data Fig. 2 Optimization of re-embedding strategy.
a, Mechanism of cDNA crosslinking using hydrogel re-embedding. Amine-modified nucleotides were incorporated into the rolling-circle amplification reaction. MA-NHS enables rapid conjugation to nucleophilic groups on the amplicons via its NHS ester under mild conditions. These functionalized methacrylamide moieties are then integrated into the hydrogel, effectively immobilizing the cDNA amplicons. b, Mechanism of cDNA crosslinking using BSPEG. Amine-modified nucleotides were incorporated into the rolling-circle amplification reaction followed by BSPEG crosslinking, where the NHS esters of BSPEG react with the amino groups on the amplicons. c, Mechanism of cDNA crosslinking using Click chemistry. Azide and alkyne groups were incorporated during the RCA process, followed by the addition of copper to catalyze the azide-alkyne cycloaddition, forming a stable triazole ring as a crosslinking method. d, Representative fluorescent imaging demonstrating sequencing signal-to-noise ratio using different cDNA crosslinking strategies. BSPEG and Click chemistry crosslinking result in higher background noise compared to hydrogel re-embedding after several rounds of sequencing. Additionally, the incorporation of azide and alkyne moieties during RCA significantly reduced amplification efficiency, leading to fewer amplicons. SNR is defined as the ratio of signal intensity to background intensity. n = 4 biological replicates per condition. Data presented as mean ± standard deviation.
Extended Data Fig. 3 Detection specificity, depth-dependent transcript quantification, and sequencing quality assessment in Deep-STARmap and Deep-RIBOmap.
a, Total read counts for 4 cell-type markers (Supplementary Table 1) as a function of tissue depth in 260-µm-thick mouse brain slices processed with Deep-STARmap (left) and Deep-RIBOmap (right). Read counts remain stable within the first 200 µm but progressively decline beyond this depth, dropping below ~75% of the initial counts at 250 µm. b, Average reads per cell for the same 4 cell-type markers as a function of tissue depth in Deep-STARmap (left) and Deep-RIBOmap (right). c, Representative fluorescent images showing DNA amplicons from 4 cell-type markers (Supplementary Table 1) (red) and nuclei (DAPI, blue) in Deep-STARmap and Deep-RIBOmap under different control conditions. Scale bar: 30 µm. d, Quantification of reads per cell for each condition, demonstrating significantly reduced signal in the absence of required probe components. Inset panels show zoomed-in views of low-signal conditions. Statistical analysis was performed using a two-sided t-test, with ***P < 0.001 and ****P < 0.0001. Data are presented as mean ± standard deviation from n = 4 technical replicates per condition. e, Representative fluorescence images of RNA amplicons (red) and nuclei (DAPI, blue) under various control conditions using four neuron markers (Supplementary Table 1). f, Quantification of reads per cell across all conditions. Bars represent mean ± s.d.; n = 4 technical replicates per condition. ***P < 0.001, two-sided t-test. Scale bar, 30 µm. g, Proportion of reads filtered due to low-quality scores across z-depth. h, Proportion of reads filtered out due to sequencing errors (not CXXXXXC) across z-depth. i, False positive rate estimation using 7 unused barcodes. The detected false positive rate (blue line) remains significantly below the theoretical threshold (red dashed line) across Z-depth.
Extended Data Fig. 4 Single-cell spatial transcriptomic and translatomic profiling of 1,017 genes in the mouse brain.
a, Dot plot illustrating the expression levels of representative markers across various major cell types. The color scale represents the log2 fold change in gene expression compared to the mean gene expression values across all cells. The dot size indicates the percentage of cells expressing the genes within each major cell type. b, Deep-STARmap (left) and Deep-RIBOmap (right) images of example cell marker genes and neurotransmitter genes. xyz size: 4.5 mm, 4.5 mm, 150 µm.
Extended Data Fig. 5 Marker gene expression and cross-modal correlation analysis.
a, Dot plot showing marker gene expression across astrocytes (AC), oligodendrocytes (OLG), di- and mesencephalon excitatory neurons (DEGLU, HABCHO, HABGLU, MEGLU), peptidergic neurons (INH), telencephalon projecting inhibitory neurons (MSN), and ependymal cells (EPEN) in thick and thin sections. The color scale represents the log2 fold change in gene expression compared to the mean gene expression values across all cells. The dot size indicates the percentage of cells expressing the genes within each major cell type. b. Pearson correlation heatmaps comparing gene expression profiles across modalities. Left two panels: correlation between Deep-STARmap or Deep-RIBOmap and thin-section STARmap. Right two panels: correlation between Deep-STARmap or Deep-RIBOmap and reference scRNA-seq profiles. Each matrix shows cell-type-level average gene expression correlation. c. Scatter plots comparing average gene expression levels between spatial methods and bulk RNA-seq from mouse cortex. Left two panels: correlation of gene expression between Deep-STARmap or Deep-RIBOmap and thin tissue STARmap. Right two panels: correlation with bulk RNA-seq. Each dot represents one gene. Gene expression values are log2-transformed and normalized. Pearson correlation coefficients (r) are reported in each plot; red dashed lines indicate the identity line.
Extended Data Fig. 6 Spatial organization of fine-grained neuronal, oligodendrocyte, and astrocyte clusters in thick and thin tissue sections.
a, Spatial distribution of TEGLU (Telencephalon projecting excitatory neurons), OLG (oligodendrocyte), AC (astrocyte), DEGLU (Di- and mesencephalon excitatory neurons), and INH (Di- and mesencephalon inhibitory neurons) subtypes in coronal mouse brain sections, shown for both thick and thin tissue preparations.
Extended Data Fig. 7 Reproducibility of Deep-STARmap and Deep-RIBOmap, spatial translatome-transcriptome comparison.
a, 3D molecular cell-type maps of two independent biological replicates for Deep-STARmap (left) and Deep-RIBOmap (right) in 150-µm-thick coronal mouse brain sections. Each dot represents an individual cell, colored by its assigned cell type. b, Dot plot visualization of gene expression profiles across major cell types in batch 2. The color scale represents the log2 fold change in gene expression compared to the mean gene expression values across all cells. The dot size indicates the percentage of cells expressing the genes within each major cell type. c, Heat map showing the gene clustering using the RIBOmap and STARmap results by cell type (Z-score expression). d, Visualization of enriched GO terms within each gene module, categorized and color-coded by module. In the enrichment map, nodes represent enriched GO terms, with the size of each node reflecting the number of genes associated with that term. Edges between nodes indicate shared genes among the GO terms. e, Heat map displaying gene clustering based on Deep-STARmap and Deep-RIBOmap results across the three oligodendrocyte lineage cell types (left). The right panel shows the relative translational efficiency (RTE) of these genes within each oligodendrocyte lineage cell type (Z-score expression). Statistical significance was assessed using pairwise Mann-Whitney U tests with Benjamini-Hochberg FDR correction. Sample sizes: gene module 1 (n = 18), Gene module 2 (n = 30), Gene module 3 (n = 26).
Extended Data Fig. 8 Localized translation in the somata and processes of neuronal and glial cells in the mouse brain.
a, Schematic illustration of a hippocampal slice highlighting the somata and processes of hippocampal neurons. b, Processes read percentages of individual translating genes with genes rank-ordered based on their process reads percentage. c, Significantly enriched GO terms for processes-enriched and somata-enriched translating genes. d, Spatial translation map of representative genes with enriched translation in processes (top) and somata (bottom) within the hippocampus, depicting somata reads in blue and process reads in red. e, f, Nearest-neighbor distance distributions from SST, PV, VIP, and LAMP5 neurons to cells of the same subtype (‘to self’) and to other inhibitory subtypes (‘to other’) in a single Deep-STARmap sample (SST: n = 3,027; PV: n = 3,280; LAMP5: n = 1,985; VIP: n = 1,073). Center line, median; box bounds, interquartile range (IQR: 25th to 75th percentile); whiskers, 1.5×IQR; outliers excluded. Two-sided independent t-test, ****P < 0.0001. g, Nearest-neighbor distance distributions in 15-µm thin sections subsampled from the same 3D Deep-STARmap volume. Binwidth = 3 µm. The y-axis shows probability density, where the area under the entire histogram equals 1.
Extended Data Fig. 9 Quantification of Tebow-labeled neurons.
a, Another zoom-in view of volume rendering of mouse cortical pyramidal neurons labeled with Tetbow. b, c, Cell count quantification of Tebow-labeled neurons across major cell types (b) and subtypes (c). d, Confusion matrix showing the performance of a classifier trained on morphological features of reconstructed neurons. The classifier distinguishes excitatory neurons, Pv interneurons, and Sst interneurons with high accuracy. The y-axis represents true neuronal subtypes, and the x-axis indicates predicted labels. Values in each cell correspond to the raw number of neurons assigned to each category.
Extended Data Fig. 10 3D spatial mapping of molecular regions, cell types, gene expression, and cell–cell adjacency analysis in cSCC.
a, Molecular tissue region maps visualized in 3D. Each dot represents a cell. Three molecular regions can be identified: tumor, fibroblast-rich stroma, and normal skin. These regions were identified by analyzing low-frequency, large-scale patterns within the spatial neighbors graph. b, 3D spatial distribution of cell types in two independent cSCC tumor sections from the same patient, mapped using Deep-STARmap. c, Gene expression profiles of marker genes across major cell types in the replicate. The color scale represents the log2 fold change in gene expression compared to the mean gene expression values across all cells. The dot size indicates the percentage of cells expressing the genes within each major cell type. d, 2D cell–cell adjacency quantified in 4 different 15 µm subslices by log enrichment, calculated as the logarithm of the ratio of observed adjacency frequency to the mean adjacency frequency from 1,000 permutations. The 2D analysis is performed by projecting 15 μm (~1 cell layer) slices along the z-axis, taken within the same 3D volume as shown in Fig. 4. e, f, The 3D analysis detects stronger patterns of cell–cell adjacencies because the number of connected cells (edges of a given cell in the mesh graph via Delaunay triangulation) is greater than in 2D. The 2D nearest-neighbor distances cannot accurately represent the 3D cellular environment.
Supplementary information
Supplementary Tables
Supplementary Tables 1–8.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sui, X., Lo, J.A., Luo, S. et al. Scalable spatial single-cell transcriptomics and translatomics in 3D thick tissue blocks. Nat Methods 22, 2574–2584 (2025). https://doi.org/10.1038/s41592-025-02867-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02867-0
This article is cited by
-
Year in review 2025
Nature Methods (2026)
-
Bridging the dimensional gap from planar spatial transcriptomics to 3D cell atlases
Nature Methods (2025)
-
From slices to deep dishes: spatial transcriptomics and translatomics of thick tissue blocks
Nature Methods (2025)