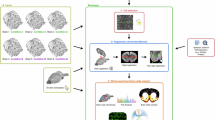
Abstract
We propose a latent space-based statistical network analysis (LatentSNA) method that implements network science in a generative Bayesian framework, preserves neurologically meaningful brain topology and improves statistical power for imaging biomarker detection. LatentSNA (1) addresses the lack of power and inflated type II errors in current analytic approaches when detecting imaging biomarkers, (2) allows unbiased estimation of the influence of biomarkers on behavioral variants, (3) quantifies uncertainty and evaluates the likelihood of estimated biomarker effects against chance and (4) improves brain–behavior prediction in new samples as well as the clinical utility of neuroimaging findings. LatentSNA is broadly applicable across multiple imaging modalities and outcome measures in developing, aging and transdiagnostic cohorts, totaling 8,003 to 11,861 participants. LatentSNA achieves substantial accuracy gains (averaging 110–150%) and replicability improvements (averaging 153%) over existing approaches in moderate to large datasets. As a result, LatentSNA elucidates how network topology is implicated in brain–behavior relationships.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The individual-level imaging and behavior data used in the present study are available from four publicly accessible data resources: ABCD (https://abcdstudy.org/), HCP (https://www.humanconnectome.org/), A4 (https://www.a4studydata.org) and ADNI (https://adni.loni.usc.edu). Transdiagnostic data are available via the NDA website (https://nda.nih.gov/).
Code availability
The estimation algorithm for this paper was implemented in R. The code is available at https://github.com/selenashuowang/latentSNA with a tutorial. The code is released under the MIT License. In this GitHub repository, we have provided instructions for installation (specifying prerequisite packages), explanations of outputs and a sample toy example with evaluations.
References
Barabási, A. Network science. Philos. Trans. A Math. Phys. Eng Sci 371, 20120375 (2013).
Sweet, T. & Wang, S. Network science in psychology. Psychol. Methods https://doi.org/10.1037/met0000745 (2025).
Wang, S. et al. Establishing group-level brain structural connectivity incorporating anatomical knowledge under latent space modeling. Med. Image Anal. 99, 103309 (2025).
Satterthwaite, T. D. et al. Common and dissociable dysfunction of the reward system in bipolar and unipolar depression. Neuropsychopharmacology 40, 2258–2268 (2015).
Shen, X. et al. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat. Protoc. 12, 506–518 (2017).
Gao, S., Greene, A. S., Constable, R. T. & Scheinost, D. Combining multiple connectomes improves predictive modeling of phenotypic measures. Neuroimage 201, 116038 (2019).
Hearst, M. A., Dumais, S. T., Osuna, E., Platt, J. & Scholkopf, B. Support vector machines. IEEE Intell. Syst. 13, 18–28 (1998).
Belgiu, M. & Drăguţ, L. Random forest in remote sensing: a review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 114, 24–31 (2016).
Albawi, S., Mohammed, T. A. & Al-Zawi, S. Understanding of a convolutional neural network. In 2017 International Conference on Engineering and Technology (ICET) 1–6 (IEEE, 2017).
Bullmore, E. & Sporns, O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198 (2009).
Dobra, A., Hans, C., Jones, B., Nevins, J. R., Yao, G. & West, M. Sparse graphical models for exploring gene expression data. J. Multivar. Anal. 90, 196–212 (2004).
Friedman, N., Linial, M., Nachman, I. & Pe’er, D. Using Bayesian networks to analyze expression data. In Proc. 4th Annual International Conference on Computational Molecular Biology 127–135 (ACM, 2000).
Ni, Y., Baladandayuthapani, V., Vannucci, M. & Stingo, F. C. Bayesian graphical models for modern biological applications. Stat. Methods Appt. 31, 197–225 (2022).
Ni, Y., Stingo, F. C. & Baladandayuthapani, V. Bayesian graphical regression. J. Am. Stat. Assoc. 114, 184–197 (2019).
Wang, S. Recent integrations of latent variable network modeling with psychometric models. Front. Psychol. 12, 773289 (2021).
Wang, S. & Edgerton, J. Resilience to stress in bipartite networks: application to the Islamic State recruitment network. J. Complex Netw. 10, cnac017 (2022).
Di Fatta, D., Caputo, F., Evangelista, F. & Dominici, G. Small world theory and the World Wide Web: linking small world properties and website centrality. Int. J. Mark. Bus. Syst. 2, 126–140 (2016).
Bassett, D. S. & Bullmore, E. D. Small-world brain networks. Neuroscientist 12, 512–523 (2006).
Petersen, R. C. et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI) clinical characterization. Neurology 74, 201–209 (2010).
Sperling, R. A. et al. The A4 study: stopping AD before symptoms begin?. Sci. Transl. Med. 6, 228fs13 (2014).
Bookheimer, S. Y. et al. The Lifespan Human Connectome Project in Aging: an overview. Neuroimage 185, 335–348 (2019).
Casey, B. J. et al. The Adolescent Brain Cognitive Development (ABCD) study: imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 32, 43–54 (2018).
Greene, A. S. et al. Brain–phenotype models fail for individuals who defy sample stereotypes. Nature 609, 109–118 (2022).
Relión, J. D. A., Kessler, D., Levina, E. & Taylor, S. F. Network classification with applications to brain connectomics. Ann. Appl. Stat. 13, 1648–1677 (2019).
Zhang, Z., Allen, G. I., Zhu, H. & Dunson, D. Tensor network factorizations: relationships between brain structural connectomes and traits. Neuroimage 197, 330–343 (2019).
Krueger, R. F. & Markon, K. E. Understanding psychopathology: melding behavior genetics, personality, and quantitative psychology to develop an empirically based model. Curr. Dir. Psychol. Sci. 15, 113–117 (2006).
Sawai, H. Exploring a new small-world network for real-world applications. In Networked Digital Technologies: 4th International Conference, NDT 2012, Dubai, UAE, April 24–26, 2012. Proceedings, Part I 4 (ed. Benlamri, R.) 90–101 (Springer, 2012).
Zhao, Y., Li, T. & Zhu, H. Bayesian sparse heritability analysis with high-dimensional neuroimaging phenotypes. Biostatistics 23, 467–484 (2022).
Zhang, X., Hulvershorn, L. A., Constable, T., Zhao, Y. & Wang, S. Cost efficiency of fMRI studies using resting-state vs. task-based functional connectivity. Hum. Brain Mapp. 46, e70260 (2025).
Chen, S. et al. Identifying covariate-related subnetworks for whole-brain connectome analysis. Biostatistics 25, 541–558 (2024).
Kolb, B. & Whishaw, I. Q. Brain plasticity and behavior. Annu. Rev. Psychol. 49, 43–64 (1998).
Chahal, R., Gotlib, I. H. & Guyer, A. E. Research review: brain network connectivity and the heterogeneity of depression in adolescence—a precision mental health perspective. J. Child Psychol. Psychiatry 61, 1282–1298 (2020).
Breiman, L. Statistical modeling: the two cultures (with comments and a rejoinder by the author). Stat. Sci. 16, 199–231 (2001).
Wang, S., Wu, H. & Pek, J. Performance of alternative regression weights in the context of prediction versus inference. Multivar. Behav. Res. 57, 163 (2022).
Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. B Stat. Methodol. 58, 267–288 (1996).
Wasserman, S. et al. Social Network Analysis: Methods and Applications (Cambridge Univ. Press, 1994).
Heider, F. Social perception and phenomenal causality. Psychol. Rev. 51, 358–374 (1944).
Hoff, P. D. Bilinear mixed-effects models for dyadic data. J. Am. Stat. Assoc. 100, 286–295 (2005).
Hoff, P. D. Modeling homophily and stochastic equivalence in symmetric relational data. Adv. Neural Inf. Process. Syst. 20, 657–664 (2008).
Sosa, J. & Buitrago, L. A review of latent space models for social networks. Rev. Colomb. Estad. 44, 171–200 (2021).
Calhoun, V. D. & Sui, J. Multimodal fusion of brain imaging data: a key to finding the missing link(s) in complex mental illness. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 1, 230–244 (2016).
Wu, J., Li, J., Eickhoff, S. B., Scheinost, D. & Genon, S. The challenges and prospects of brain-based prediction of behaviour. Nat. Hum. Behav. 7, 1255–1264 (2023).
Stavropoulos, I. et al. Increased hair cortisol and antecedent somatic complaints in children with a first epileptic seizure. Epilepsy Behav. 68, 146–152 (2017).
Shen, X., Papademetris, X. & Constable, R. T. Graph-theory based parcellation of functional subunits in the brain from resting-state fMRI data. Neuroimage 50, 1027–1035 (2010).
Gelman, A., Carlin, J. B., Stern, H. S. & Rubin, D. B. Bayesian Data Analysis (Chapman and Hall/CRC, 1995).
Price, J. L. & Drevets, W. C. Neural circuits underlying the pathophysiology of mood disorders. Trends Cogn. Sci. 16, 61–71 (2012).
Chen, J. et al. Shared and unique brain network features predict cognitive, personality, and mental health scores in the ABCD study. Nat. Commun. 13, 2217 (2022).
Cole, M. W., Bassett, D. S., Power, J. D., Braver, T. S. & Petersen, S. E. Intrinsic and task-evoked network architectures of the human brain. Neuron 83, 238–251 (2014).
Guell, X., Gabrieli, J. D. E. & Schmahmann, J. D. Triple representation of language, working memory, social and emotion processing in the cerebellum: convergent evidence from task and seed-based resting-state fMRI analyses in a single large cohort. Neuroimage 172, 437–449 (2018).
Liu, Z. et al. Decreased regional homogeneity in insula and cerebellum: a resting-state fMRI study in patients with major depression and subjects at high risk for major depression. Psychiatry Res. Neuroimaging 182, 211–215 (2010).
Zhuo, C. et al. Altered resting-state functional connectivity of the cerebellum in schizophrenia. Brain Imaging Behav. 12, 383–389 (2018).
D’Agostino, R. B. Transformation to normality of the null distribution of g1. Biometrika 57, 679–681 (1970).
Anscombe, F. J. & Glynn, W. J. Distribution of the kurtosis statistic b2 for normal samples. Biometrika 70, 227–234 (1983).
Nigam, S. et al. Rich-club organization in effective connectivity among cortical neurons. J. Neurosci. 36, 670–684 (2016).
Guimerà, R., Díaz-Guilera, A., Vega-Redondo, F., Cabrales, A. & Arenas, A. Optimal network topologies for local search with congestion. Phys. Rev. Lett. 89, 248701 (2002).
Madore, K. P. & Wagner, A. D. Multicosts of multitasking. Cerebrum 2019, cer-04-19 (2019).
Gollini, I. & Murphy, T. B. Joint modeling of multiple network views. J. Comput. Graph. Stat. 25, 246–265 (2016).
Salter-Townshend, M. & McCormick, T. H. Latent space models for multiview network data. Ann. Appl. Stat. 11, 1217–1244 (2017).
Wang, L., Durante, D., Jung, R. E. & Dunson, D. B. Bayesian network–response regression. Bioinformatics 33, 1859–1866 (2017).
Morris, E. L., He, K. & Kang, J. Scalar on network regression via boosting. Ann. Appl. Stat. 16, 2755–2773 (2022).
Fischer, G. H. & Molenaar, I. W. (eds) Rasch Models: Foundations, Recent Developments, and Applications (Springer, 2012).
Joshi, A. et al. Unified framework for development, deployment and robust testing of neuroimaging algorithms. Neuroinformatics 9, 69–84 (2011).
Greene, A. S., Gao, S., Scheinost, D. & Constable, R. T. Task-induced brain state manipulation improves prediction of individual traits. Nat. Commun. 9, 2807 (2018).
Horien, C., Shen, X., Scheinost, D. & Constable, R. T. The individual functional connectome is unique and stable over months to years. Neuroimage 189, 676–687 (2019).
Shen, X., Tokoglu, F., Papademetris, X. & Constable, R. T. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage 82, 403–415 (2013).
Manjón, J. V. et al. Diffusion weighted image denoising using overcomplete local PCA. PLoS ONE 8, e73021 (2013).
Moore, C. & Sciacca, F. Fiber assignment by continuous tracking algorithm (FACT). Radiopaedia https://doi.org/10.53347/rid-72014 (2019).
Jenkinson, M., Beckmann, C. F., Behrens, T. E. J., Woolrich, M. W. & Smith, S. M. FSL. Neuroimage 62, 782–790 (2012).
Cammoun, L. et al. Mapping the human connectome at multiple scales with diffusion spectrum MRI. J. Neurosci. Methods 203, 386–397 (2012).
Yan, J. et al. Brain-wide structural connectivity alterations under the control of Alzheimer risk genes. Int. J. Comput. Biol. Drug Des. 13, 58–70 (2020).
Tourbier, S. et al. Connectome Mapper 3: a flexible and open-source pipeline software for multiscale multimodal human connectome mapping. J. Open Source Softw. 7, 4248 (2022).
Gorgolewski, K. et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in Python. Front. Neuroinform. 5, 13 (2011).
Cox, R. W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29, 162–173 (1996).
Jenkinson, M. & Smith, S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 5, 143–156 (2001).
Kueper, J. K., Speechley, M. & Montero-Odasso, M. The Alzheimer’s Disease Assessment Scale–Cognitive Subscale (ADAS-Cog): modifications and responsiveness in pre-dementia populations. a narrative review. J. Alzheimers Dis. 63, 423–444 (2018).
Farias, S. T. et al. The measurement of everyday cognition (ECog): scale development and psychometric properties. Neuropsychology 22, 531–544 (2008).
Lutkenhoff, E. S. et al. Optimized brain extraction for pathological brains (optiBET). PloS ONE 9, e115551 (2014).
Greve, D. N. et al. Cortical surface-based analysis reduces bias and variance in kinetic modeling of brain PET data. Neuroimage 92, 225–236 (2014).
Desikan, R. S. et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980 (2006).
Donohue, M. C. et al. The Preclinical Alzheimer Cognitive Composite: measuring amyloid-related decline. JAMA Neurol. 71, 961–970 (2014).
Papp, K. V. et al. The Computerized Cognitive Composite (c3) in A4, an Alzheimer’s disease secondary prevention trial. J. Prev. Alzheimers Dis. 8, 59–67 (2021).
Weintraub, S. et al. Cognition assessment using the NIH Toolbox. Neurology 80, S54–S64 (2013).
NIH Toolbox Scoring and Interpretation Guide (NIH, 2022); https://www.epicrehab.com/epic/documents/crc/crc-201307-nih-toolbox-scoring-and-interpretation-manual%209-27-12.pdf
Derogatis, L. R. & Melisaratos, N. The brief symptom inventory: an introductory report. Psychol. Med. 13, 595–605 (1983).
Zhang, J., Sun, W. W. & Li, L. Generalized connectivity matrix response regression with applications in brain connectivity studies. J. Comput. Graph. Stat. 32, 252–262 (2023).
Wang, S., Paul, S. & De Boeck, P. Joint latent space model for social networks with multivariate attributes. Psychometrika 88, 1197–1227 (2023).
Leonard, T. & Hsu, J. S. J. Bayesian inference for a covariance matrix. Ann. Stat. 20, 1669–1696 (1992).
Zhang, Z. A note on Wishart and inverse Wishart priors for covariance matrix. J. Behav. Data Sci. 1, 119–126 (2021).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010).
González, I., Déjean, S., Martin, P. G. P. & Baccini, A. CCA: an R package to extend canonical correlation analysis. J. Stat. Softw. 23, 1–14 (2008).
Aine, C. et al. Multimodal neuroimaging in schizophrenia: description and dissemination. Neuroinformatics 15, 343–364 (2017).
Meyer, D., Dimitriadou, E., Hornik, K., Weingessel, A. & Leisch, F. e1071: misc functions of the Department of Statistics, Probability Theory Group (formerly: E1071), TU Wien. https://CRAN.R-project.org/package=e1071 (2023).
Wright, M. N. & Ziegler, A. ranger: a fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 77, 1–17 (2017).
Falbel, D. & Luraschi, J. torch: tensors and neural networks with ‘GPU’ acceleration. https://CRAN.R-project.org/package=torch (2023).
Achard, S., Salvador, R., Whitcher, B., Suckling, J. & Bullmore, E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J. Neurosci. 26, 63–72 (2006).
Paquette, M., Girard, G., Chamberland, M. & Descoteaux, M. ‘Noise’ in diffusion tractography connectomes is not additive. In ISMRM 24th Annual Meeting (ISMR, 2016).
Newman, M. E. J. Scientific collaboration networks. II. Shortest paths, weighted networks, and centrality. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 64, 016132 (2001).
Freeman, L. C. Centrality in social networks: conceptual clarification. Soc. Netw. 1, 238–263 (1979).
Freeman, L. C. A set of measures of centrality based on betweenness. Sociometry 40, 35–41 (1977).
Brandes, U. A faster algorithm for betweenness centrality. J. Math. Sociol. 25, 163–177 (2001).
Burt, R. S. in The New Economic Sociology: A Reader (ed. Dobbin, F.) 325–348 (Princeton Univ. Press, 2004).
Acknowledgements
S.W. was partially supported by the Alzheimer’s Disease Data Initiative from the 2025 William H. Gates Sr Fellowship and NIH grant P30 AG072976. Y.Z. was partially supported by NIH grants R01AG068191, RF1AG081413, R01EB034720, P30AG072976 and P30AG021342. We thank the individuals represented in the ADNI (https://adni.loni.usc.edu), A4 (https://www.a4studydata.org), ABCD (https://nda.nih.gov/abcd), HCP (https://db.humanconnectome.org) and transdiagnostic studies for their participation and the research teams for their work in collecting, processing and disseminating these datasets for analysis. Some data collection and sharing for this project was funded by the ADNI (NIH grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). The A4 study is a secondary prevention trial in preclinical Alzheimer’s disease, aiming to slow cognitive decline associated with brain Aβ accumulation in clinically typical older individuals. Some data used in the preparation of this article were obtained from the ABCD study, held in the National Institute of Mental Health Data Archive (https://nda.nih.gov). HCP data were provided by the HCP WU-Minn Consortium (principal investigators D. Van Essen and K. Ugurbil; 1U54MH091657) funded by the 16 NIH institutes and centers that support the NIH Blueprint for Neuroscience Research and the McDonnell Center for Systems Neuroscience at Washington University. The transdiagnostic study was supported by the National Institute of Mental Health (R01MH123245 and R01MH120080).
Author information
Authors and Affiliations
Contributions
S.W. and Y.Z. designed the study; S.W. and Y.L. performed the simulation; S.W., W.X., X.T. and X.Z. analyzed data; S.W. wrote the paper; and all authors have given comments and edits. All authors contributed to the interpretation of results, discussion and paper revision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Chao Huang, Pew-Thian Yap and Aiying Zhang for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Nina Vogt, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
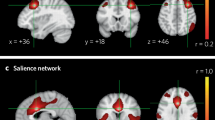
Extended Data Fig. 1 Functional architectures of internalizing psychopathology are driven by the core actors of the connectivity network.
(A) The strength of each brain region in MOT based on the latent network (left) and the observed network (right) for an average participant during MID condition. Regions identified to play a significant role in explaining individual differences in internalizing behaviors are colored as green, and non-significant regions are colored as red. (B) The location and connectivity networks of an imaging biomarker (top) versus a null effect (bottom) with no identifiable contribution to internalizing. The 3D brain plots show the front (top left), back (top right), right (bottom left) and left (bottom right) views. (C) The circle plots of the connectivity edges associated with the imaging biomarker (top) and the null effect (bottom). In B and C, red line indicates positive connectivity edges, and blue line indicates negative connectivity edges.
Extended Data Fig. 2 Internalizing psychopathology in children are attributable to star-like functional networks.
Latent internalizing networks (left) against CPM networks (right) for an average participant in each functional system during MID task. Node positions of the latent networks are then determined using the fruchterman-reingold force-directed graph layout algorithm. The nodes are fixed in the same positions when plotting the internalizing connectivity edges identified via CPM. We also show the corresponding (whole) latent networks, with both significant and non-significant connectivity edges, estimated via LatentSNA, as well as the average observed networks. MF: Medial-Frontal, FP: Fronto-parietal, DMN: Default Mode, MOT: Motor, VI: Visual I, VII: Visual II, VAs: Visual Association, LIM: Limbic, BG: Basal Ganglia, CBL: Cerebellum. Blue represents the positive connectivity edges, and red represents negative edges.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, S., Zhang, X., Liu, Y. et al. Latent space-based network analysis for brain–behavior linking in neuroimaging. Nat Methods 23, 225–235 (2026). https://doi.org/10.1038/s41592-025-02896-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02896-9