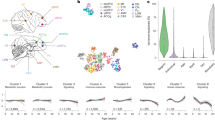
Abstract
A systematic investigation of aging patterns across virtually all major tissues in nonhuman primates, our evolutionarily closest relatives, can provide valuable insights into tissue aging in humans, which is still elusive largely due to the difficulty in sampling. Here, we generated and analyzed multi-omics data, including transcriptome, proteome and metabolome, from 30 tissues of 17 female rhesus macaques (Macaca mulatta) aged 3–27 years. We found that certain molecular features, such as increased inflammation, are consistent across tissues and align with findings in mice and humans. We further revealed that tissue aging in macaques is asynchronous and can be classified into two distinct types, with one type exhibiting more pronounced aging degree, likely associated with decreased mRNA translation efficiency, and predominantly contributing to whole-body aging. This work provides a comprehensive molecular landscape of aging in nonhuman primate tissues and links translation efficiency to tissue-specific aging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The omics data are freely and publicly available. Level-2 data, including transcriptome raw counts and log2 peak areas for the proteome and metabolome, are accessible on Figshare at https://doi.org/10.6084/m9.figshare.26963386 (ref. 64). For the level-1 raw data, the raw FASTQ files of the transcriptome have been deposited in the Genome Sequence Archive at the National Genomics Data Center65,66 (GSA CRA026248), publicly accessible at https://ngdc.cncb.ac.cn/gsa. The raw LC–MS/MS data for the proteome have been deposited to the ProteomeXchange Consortium via the PRIDE67 partner repository with the dataset identifier PXD066108 and also deposited to OMIX66 (https://ngdc.cncb.ac.cn/omix, accession no. OMIX001778). The raw LC–MS/MS data for the metabolome are deposited in OMIX (https://ngdc.cncb.ac.cn/omix, accession no. OMIX001779).
Code availability
The code to reproduce the results in this study is available on Figshare at https://doi.org/10.6084/m9.figshare.26963386 (ref. 64) or GitHub at https://github.com/GonghuaLi/Macaca_30tissue_aging. Supplementary Data 10 provides a comprehensive PDF document generated from the coding of R Markdown (RMD) file, containing all code and results.
References
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Kaeberlein, M., Rabinovitch, P. S. & Martin, G. M. Healthy aging: the ultimate preventative medicine. Science 350, 1191–1193 (2015).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell 186, 243–278 (2023).
Schaum, N. et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nature 583, 596–602 (2020).
Rando, T. A. & Wyss-Coray, T. Asynchronous, contagious and digital aging. Nat. Aging 1, 29–35 (2021).
Zhang, W. et al. SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 560, 661–665 (2018).
Colman, R. J. Non-human primates as a model for aging. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 2733–2741 (2018).
Anderson, R. M. & Colman, R. J. Prospects and perspectives in primate aging research. Antioxid. Redox Signal. 14, 203–205 (2011).
Warren, W. C. et al. Sequence diversity analyses of an improved rhesus macaque genome enhance its biomedical utility. Science 370, eabc6617 (2020).
Wang, S. et al. Single-cell transcriptomic atlas of primate ovarian aging. Cell 180, 585–600.e519 (2020).
Liang, D. et al. Cellular and molecular landscape of mammalian sinoatrial node revealed by single-cell RNA sequencing. Nat. Commun. 12, 287 (2021).
Zhang, W. et al. A single-cell transcriptomic landscape of primate arterial aging. Nat. Commun. 11, 2202 (2020).
Li, M.-L. et al. 547 transcriptomes from 44 brain areas reveal features of the aging brain in nonhuman primates. Genome Biol. 20, 258 (2019).
Wang, S. et al. Deciphering primate retinal aging at single-cell resolution. Protein Cell 12, 889–898 (2021).
Huang, D. et al. A single-nucleus transcriptomic atlas of primate testicular aging reveals exhaustion of the spermatogonial stem cell reservoir and loss of Sertoli cell homeostasis. Protein Cell 14, 888–907 (2023).
Li, J. et al. A single-cell transcriptomic atlas of primate pancreatic islet aging. Natl Sci. Rev. 8, nwaa127 (2021).
Jing, Y. et al. Single-nucleus profiling unveils a geroprotective role of the FOXO3 in primate skeletal muscle aging. Protein Cell 14, 497–512 (2023).
Wolters, S. & Schumacher, B. Genome maintenance and transcription integrity in aging and disease. Front. Genet. 4, e19 (2013).
Zierer, J., Menni, C., Kastenmüller, G. & Spector, T. D. Integration of ‘omics’ data in aging research: from biomarkers to systems biology. Aging Cell 14, 933–944 (2015).
Consortium, G. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Melé, M. et al. The human transcriptome across tissues and individuals. Science 348, 660–665 (2015).
Jiang, L. et al. A quantitative proteome map of the human body. Cell 183, 269–283.e219 (2020).
Palmer, D., Fabris, F., Doherty, A., Freitas, A. A. & de Magalhães, J. P. Ageing transcriptome meta-analysis reveals similarities and differences between key mammalian tissues. Aging 13, 3313 (2021).
Rozowsky, J. et al. The EN-TEx resource of multi-tissue personal epigenomes & variant-impact models. Cell 186, 1493–1511.e40 (2023).
Keele, G. R. et al. Global and tissue-specific aging effects on murine proteomes. Cell Rep. 42, 112715 (2023).
Shen, X. et al. Nonlinear dynamics of multi-omics profiles during human aging. Nat. Aging 4, 1619–1634 (2024).
Contrepois, K. et al. Molecular choreography of acute exercise. Cell 181, 1112–1130.e1 (2020).
Kumar, L. & Futschik, M. E. Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2, 5 (2007).
Avelar, R. A. et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 21, 91 (2020).
Battle, A. et al. Impact of regulatory variation from RNA to protein. Science 347, 664–667 (2015).
Chick, J. M. et al. Defining the consequences of genetic variation on a proteome-wide scale. Nature 534, 500–505 (2016).
Kim, M.-S. et al. A draft map of the human proteome. Nature 509, 575–581 (2014).
Li, G. H. et al. System-level metabolic modeling facilitates unveiling metabolic signature in exceptional longevity. Aging cell 21, e13595 (2022).
Aging Atlas, C. Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res. 49, D825–D830 (2021).
Huang, K. et al. AgeAnnoMO: a knowledgebase of multi-omics annotation for animal aging. Nucleic Acids Res. 52, D822–D834 (2024).
Widjaja, A. A. et al. Inhibition of IL-11 signalling extends mammalian healthspan and lifespan. Nature 632, 157–165 (2024).
Colman, R. J., McKiernan, S. H., Aiken, J. M. & Weindruch, R. Muscle mass loss in rhesus monkeys: age of onset. Exp. Gerontol. 40, 573–581 (2005).
Nilwik, R. et al. The decline in skeletal muscle mass with aging is mainly attributed to a reduction in type II muscle fiber size. Exp. Gerontol. 48, 492–498 (2013).
Roy, R. R. et al. Fiber size and myosin phenotypes of selected rhesus hindlimb muscles after a 14-day spaceflight. J. Gravit. Physiol. 6, 55–62 (1999).
Ohyama, Y., Redheuil, A., Kachenoura, N., Ambale Venkatesh, B. & Lima, J. A. Imaging insights on the aorta in aging. Circulation Cardiovasc. Imaging 11, e005617 (2018).
Rezzani, R., Nardo, L., Favero, G., Peroni, M. & Rodella, L. F. Thymus and aging: morphological, radiological, and functional overview. Age 36, 313–351 (2014).
Gingold, H. & Pilpel, Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 7, e481 (2011).
Tahmasebi, S., Sonenberg, N., Hershey, J. W. & Mathews, M. B. Protein synthesis and translational control: a historical perspective. Cold Spring Harb. Perspect. Biol. 11, a035584 (2019).
Wei, Y. N. et al. Transcript and protein expression decoupling reveals RNA binding proteins and miRNAs as potential modulators of human aging. Genome Biol. 16, 41 (2015).
Kelmer Sacramento, E. et al. Reduced proteasome activity in the aging brain results in ribosome stoichiometry loss and aggregation. Mol. Syst. Biol. 16, e9596 (2020).
Gerdes Gyuricza, I. et al. Genome-wide transcript and protein analysis highlights the role of protein homeostasis in the aging mouse heart. Genome Res. 32, 838–852 (2022).
Hu, Z. et al. Ssd1 and Gcn2 suppress global translation efficiency in replicatively aged yeast while their activation extends lifespan. eLife 7, e35551 (2018).
Syntichaki, P., Troulinaki, K. & Tavernarakis, N. Protein synthesis is a novel determinant of aging in Caenorhabditis elegans. Ann. NY Acad. Sci. 1119, 289–295 (2007).
Gerashchenko, M. V., Peterfi, Z., Yim, S. H. & Gladyshev, V. N. Translation elongation rate varies among organs and decreases with age. Nucleic Acids Res. 49, e9 (2021).
Yu, L. et al. IgG is an aging factor that drives adipose tissue fibrosis and metabolic decline. Cell Metab. 36, 793–807.e795 (2024).
de Magalhaes, J. P., Curado, J. & Church, G. M. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25, 875–881 (2009).
Liu, X. et al. Clusterin transduces Alzheimer-risk signals to amyloidogenesis. Signal Transduct. Target. Ther. 7, 325 (2022).
Perez-Jimenez, M. M. et al. Steroid hormones sulfatase inactivation extends lifespan and ameliorates age-related diseases. Nat. Commun. 12, 49 (2021).
Benonisson, H. et al. CD3-bispecific antibody therapy turns solid tumors into inflammatory sites but does not install protective memory. Mol. Cancer Ther. 18, 312–322 (2019).
Horai, Y., Kakimoto, T., Takemoto, K. & Tanaka, M. Quantitative analysis of histopathological findings using image processing software. J. Toxicol. Pathol. 30, 351–358 (2017).
Vijayashree, R., Aruthra, P. & Rao, K. R. A comparison of manual and automated methods of quantitation of oestrogen/progesterone receptor expression in breast carcinoma. J. Clin. Diagn. Res. 9, EC01-05 (2015).
Wang, S., Yang, D. M., Rong, R., Zhan, X. & Xiao, G. Pathology image analysis using segmentation deep learning algorithms. Am. J. Pathol. 189, 1686–1698 (2019).
Jiao, X. et al. Design mechanism and property of the novel fluorescent probes for the identification of Microthrix parvicella in situ. Materials https://doi.org/10.3390/ma10070804 (2017).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Schroeder, A. et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7, 3 (2006).
Zhao, Y. et al. TPM, FPKM, or normalized counts? A comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J. Transl. Med. 19, 1–15 (2021).
Zhao, S., Ye, Z. & Stanton, R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA 26, 903–909 (2020).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Li, G.-H. & Kong, Q.-P. A multi-omics molecular landscape of 30 tissues in aging female rhesus macaques (Macaca mulatta). Figshare https://doi.org/10.6084/m9.figshare.26963386 (2025).
Chen, T. et al. The Genome Sequence Archive family: toward explosive data growth and diverse data types. Genomics Proteom. Bioinform. 19, 578–583 (2021).
Members, C.-N. & Partners. Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2025. Nucleic Acids Res. 53, D30–D44 (2025).
Perez-Riverol, Y. et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 53, D543–D553 (2025).
Acknowledgements
This work was supported by National Key R&D Program of China (2023YFC3603400 to Q.-P.K.), National Natural Science Foundation of China (82430049 to Q.-P.K.), the CAS Project for Young Scientists in Basic Research (YSBR-076 to Q.-P.K.), Major Special Projects of Yunnan province (2018ZF007), the project entitled ‘Transformation of sub totipotent stem cells based on the tree shrew model of multiple organ dysfunction syndrome’ (SYDW[2020]19), West Light Foundation (to F.-H.X.) of the Chinese Academy of Sciences, Yunnan Province Kunming Medical University Joint Special Key Project (202301AY070001-034), Key Technologies and Translational Research on Clinical-Grade Umbilical Cord Mesenchymal Stem Cell Products for Reversing Aging, Yunnan Applied Basic Research Project (202401AW070011, 202101AS070058, 202201AS070080 and 202301AT070281), Reserve Talent Project of Young and Middle-aged Academic and Technical Leaders in Yunnan Province (202305AC160029), High-level Talent Promotion and Training Project of Kunming (Spring City Plan; 2020SCP001 to Q.-P.K.), Yunnan Revitalization Talent Support Program Yunling Scholar Project (Q.-P.K.), Yunnan Revitalization Talent Support Program Young Talent Project (G.-H.L.), Ningbo Yongjiang Talent Introduction Programme (F.-H.X.) and Yunnan Revitalization Talent Support Program Top team (202505AT350003 and 202405AS350022). We thank Novogene for technical assistance in the transcriptomics, proteomics and metabolomics data acquisition, and C. Watts for help in proofreading the manuscript.
Author information
Authors and Affiliations
Contributions
Q.-P.K. and X.-H.P. conceived and designed the study. G.-H.L. performed the multi-omics analysis. X.-Q.Z. conducted the experimental validation. G.-H.L., F.-H.X., F.-Q.Y. and M.-X.G. performed the statistical analysis. X.-Q.Z., X.Z., L.L., L.C., Q.W. and J.Z. collected samples. X.-Q.Z., X.Z., L.C., Q.W. and J.Z. performed H&E staining and scoring. T.C., Z.L., G.R., R.P., J.G. and L.M. performed immunofluorescence and scoring. G.-H.L., F.-H.X., X.-Q.Z., X.-H.P. and Q.-P.K. drafted and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks João Pedro de Magalhães, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Madhura Mukhopadhyay, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Aging-related mRNAs and proteins across 30 tissues in macaques.
Aging-associated mRNAs and proteins overlapping across 30 tissues were identified using a false discovery rate (FDR) < 0.05 and |βp|> 0.008. Here, βp represents the age effect size derived from a pooled linear regression model (molecular abundance ~ age + tissue), while βt represents the age effect size from tissue-specific linear models (molecular abundance ~ age per tissue). For each individual tissue, statistical significance was assessed using two-sided t-tests on the age coefficients derived from the tissue-specific linear models, and these P values were not adjusted for multiple comparisons. Exact P values are provided in Supplementary Data 1-2. The P values of <0.05, <0.01, and <0.001 in each tissue are denoted by *, **, and ***, respectively.
Extended Data Fig. 2 Tissue-specific aging-related mRNAs and proteins in 30 tissues.
A linear regression model was applied to each tissue to identify aging-associated mRNAs and proteins, selecting those with P < 0.05 and an absolute age effect size (|βt|) > 0.04. To ensure tissue specificity, candidate molecules were further required to be significant (P < 0.05) in no more than two tissues. a-b, the upregulated (a) and downregulated (b) tissue-specific aging-related mRNAs. c-d, the upregulated (c) and downregulated (d) tissue-specific aging-related proteins. Top one marker for each tissue is shown to the right of the heatmap. STG: Superior_temporal_gyrus; SG: Supramarginal_gyrus.
Extended Data Fig. 3 Comparison of aging-related molecules in mice, macaques, and humans.
a-c, Volcano plots of aging-related mRNAs in mice (a), macaques (b), and humans (c). d-e, Venn plots comparing upregulated (d) and downregulated (e) mRNAs among mice, macaques, and humans. Overlapped upregulated aging-related mRNAs across the three species include B2M, ANXA1 and CSF1. Overlapped downregulated aging-related mRNAs include COL3A1, PIK3R1, and PANK1. f-g, Volcano plots of aging-related proteins in mice (f) and macaques (g). Note that the significance of protein data in mice is directly derived from Gregory’s study (Cell Reports, 2023). h, Venn plot of significantly changed aging-related proteins. Among these, 28 overlapped upregulated proteins between mice and macaques include IGHM, PZP, IGKC, ITIH4, JCHAIN, KNG1, F2, AGT and CP, while three downregulated proteins are NRAS, FKBP4 and HSPH1.
Extended Data Fig. 4 Molecular aging trajectories using proteome and metabolome.
a, Tissue-specific aging trajectories for each cluster of proteins and metabolites based on normalized molecular abundances. b, Heatmap of protein enrichment for each cluster. Statistical significance for GO enrichment was evaluated using one-sided hypergeometric tests, and P values were adjusted for multiple comparisons using the Benjamini-Hochberg method to control the false discovery rate (FDR). GO terms with adjusted P < 1×10⁻⁸ are in blue. c, Heatmap of hierarchical clustering based on tissue-specific trajectories of proteins and metabolites. d, Scatter plot of viability and molecular alteration amplitude (MAA) in 30 tissues for each cluster. MAA was calculated as the change in average Z-score of molecular abundance in each cluster between the end point (elderly group) and starting point (juvenile group). Viability was determined as the average standard deviation of the molecular trajectories within each cluster. e, Heatmap of MAA for each cluster across tissues. f, K-means consensus clustering of protein and metabolite trajectories, setting k-values from 2 to 6. g, Tissue clustering by three different methods based on the aging trajectories of proteins and metabolites.
Extended Data Fig. 5 Molecular aging trajectories using transcriptome, proteome and metabolome.
a, Whole-body aging trajectories of mRNAs based on normalized gene expression. b, Tissue-specific aging trajectories of mRNAs in 30 tissues for each cluster. c, Heatmap of hierarchical clustering based on combined tissue-specific trajectories, incorporating both mRNA trajectories and protein–metabolite trajectories. d, Heatmap of MAA for each cluster across tissues from the combined tissue-specific trajectories. e, K-means consensus clustering of aging trajectories of the combined tissue-specific trajectories with k-values ranging from 2 to 6. f, Tissue clustering by three different methods based on the aging trajectories of mRNAs, proteins and metabolites.
Extended Data Fig. 6 Molecular marker changes in Type I tissues.
a, Immunofluorescence staining of p16 and p21 in seven representative Type I tissues, including spleen, thymus, aortic arch, ovary, thyroid gland, stomach, and kidney, in juvenile, young, and elderly groups. p16 and p21 are labeled in green and red, respectively. b-c, p16 (b) and p21 (c) area density in different groups in seven representative Type I tissues. Data are presented as mean ± SD with overlaid individual data points (dot plots). n = 5 sections per group based on availability, derived from multiple sections per tissue sample (see Methods). Statistical comparisons were performed using two-sided t-tests. Exact P values are provided in Supplementary Table 1. *, **, and *** represent P values of <0.05, <0.01, and <0.001, respectively. Juv: Juvenile group; Yng: Young group; Eld: Elderly group.
Extended Data Fig. 7 Molecular marker changes in Type II tissues.
a, Immunofluorescence staining of p16 and p21 in five representative Type II tissues, including muscle, pancreas, liver, lung, and back skin in juvenile, young, and elderly groups. p16 and p21 are labeled in green and red, respectively. b-c, p16 (b) and p21 (c) area density in different groups in five representative Type II tissues. Data are presented as mean ± SD with overlaid individual data points (dot plots). n = 5 sections per group based on availability, derived from multiple sections per tissue sample (see Methods). Statistical comparisons were performed using two-sided t-tests. Exact P values are provided in Supplementary Table 1. *, **, and *** represent P values of <0.05, <0.01, and <0.001, respectively. Juv: Juvenile group; Yng: Young group; Eld: Elderly group.
Extended Data Fig. 8 Histological differences in Type I tissues.
Histological differences among juvenile, young, and elderly groups in seven representative Type I tissues (spleen, thymus, aortic arch, stomach, kidney, ovary, and thyroid gland) using hematoxylin and eosin (H&E) staining. The low-power microscope partial image (marked by the black box) is magnified on the right. Histograms on the far right depict the numbers of functional cells in each tissue for the three age groups. Data are presented as mean ± SD with overlaid individual data points (dot plots). n = 5 sections per group based on availability, derived from multiple sections per tissue sample (see Methods). Statistical comparisons were performed using two-sided t-tests. Exact P values are provided in Supplementary Table 1. *, **, and *** represent P values of <0.05, <0.01, and <0.001, respectively. Juv: Juvenile group; Yng: Young group; Eld: Elderly group.
Extended Data Fig. 9 Histological differences in Type II tissues.
Histological differences among juvenile, young, and elderly groups in five representative Type II tissues (liver, lung, muscle, back skin, and pancreas) using hematoxylin and eosin (H&E) staining. Data are presented as mean ± SD with overlaid individual data points (dot plots). n = 5 sections per group based on availability, derived from multiple sections per tissue sample (see Methods). Statistical comparisons were performed using two-sided t-tests. Exact P values are provided in Supplementary Table 1. *, **, and *** represent P values of <0.05, <0.01, and <0.001, respectively. Juv: Juvenile group; Yng: Young group; Eld: Elderly group.
Extended Data Fig. 10 Changes in mRNA translation efficiency with aging.
a, Design and algorithm for estimating mRNA translation efficiency by integration of transcriptomes and proteomes. We hypothesize that, under a steady-state, protein concentration is stable, thus protein synthesis from mRNA is equal to protein degradation from existing proteins. In other words, the protein-to-mRNA ratio equals the mRNA translation efficiency-to-protein degradation (TED) ratio. Changes in TED (cTED) with aging were calculated using a linear regression model (TED ~ age), based on its age effect size (βt). b, Distribution of cTED in different tissues. Type I, II, and undefined tissues are in tomato, blue, and gold, respectively.
Supplementary information
Supplementary Information
Supplementary Notes 1–4, Supplementary Table 1 and Supplementary Figs. 1–16.
Supplementary Data 1
Aging-associated mRNAs in each tissue.
Supplementary Data 2
Aging-associated proteins in each tissue.
Supplementary Data 3
Aging-associated metabolites in each tissue.
Supplementary Data 4
Common aging-associated mRNAs across 30 tissues.
Supplementary Data 5
Common aging-associated proteins across 30 tissues.
Supplementary Data 6
Common aging-associated metabolites across 30 tissues.
Supplementary Data 7
Tissue-specific aging-associated mRNAs for each tissue.
Supplementary Data 8
Tissue-specific aging-associated proteins for each tissue.
Supplementary Data 9
Tissue-specific aging-associated metabolites for each tissue.
Supplementary Data 10
Source code and scripts used for data analysis and figure generation, along with the corresponding outputs to reproduce the results.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, GH., Zhu, XQ., Xiao, FH. et al. A multi-omics molecular landscape of 30 tissues in aging female rhesus macaques. Nat Methods 22, 2658–2669 (2025). https://doi.org/10.1038/s41592-025-02912-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02912-y