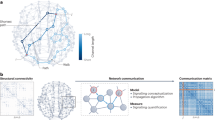
Abstract
Recent technological advances have enabled measurement of the synaptic wiring diagram, or ‘connectome’, of large neural circuits or entire brains. However, the extent to which such data constrain models of neural dynamics and function is debated. In this study, we developed a theory of connectome-constrained neural networks in which a ‘student’ network is trained to reproduce the activity of a ground truth ‘teacher’, representing a neural system for which a connectome is available. Unlike standard paradigms with unconstrained connectivity, the two networks have the same synaptic weights but different biophysical parameters, reflecting uncertainty in neuronal and synaptic properties. We found that a connectome often does not substantially constrain the dynamics of recurrent networks, illustrating the difficulty of inferring function from connectivity alone. However, recordings from a small subset of neurons can remove this degeneracy, producing dynamics in the student that agree with the teacher. Our theory demonstrates that the solution spaces of connectome-constrained and unconstrained models are qualitatively different and determines when activity in such networks can be well predicted. It can also prioritize which neurons to record to most effectively inform such predictions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The connectomics data used in this study were published in Zarin et al.16 for Drosophila larva, in Scheffer et al.9 for the central complex of adult Drosophila and in Vishwanathan et al.41 for the brainstem of the larval zebrafish. All generated data shown in the main results, together with the teacher and student recurrent networks, are publicly available at https://doi.org/10.5281/zenodo.16618353 (ref. 65).
Code availability
All simulations and analyses were performed using custom code written in Python (https://www.python.org). The code used to generate all the results and can be found in ref. 65 and https://github.com/emebeiran/connconstr.
References
Das, A. & Fiete, I. R. Systematic errors in connectivity inferred from activity in strongly recurrent networks. Nat. Neurosci. 23, 1286–1296 (2020).
Haber, A. & Schneidman, E. Learning the architectural features that predict functional similarity of neural networks. Phys. Rev. X 12, 021051 (2022).
Levina, A., Priesemann, V. & Zierenberg, J. Tackling the subsampling problem to infer collective properties from limited data. Nat. Rev. Phys. 4, 770–784 (2022).
Liang, T. & Brinkman, B. A. Statistically inferred neuronal connections in subsampled neural networks strongly correlate with spike train covariances. Phys. Rev. E 109, 044404 (2024).
Dinc, F., Shai, A., Schnitzer, M. & Tanaka, H. CORNN: convex optimization of recurrent neural networks for rapid inference of neural dynamics. Adv. Neural Inf. Process. Syst. 36, 51273–51301 (2023).
White, J. G., Southgate, E., Thomson, J. N. & Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340 (1986).
Ohyama, T. et al. A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520, 633–639 (2015).
Zheng, Z. et al. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell 174, 730–743 (2018).
Scheffer, L. K. et al. A connectome and analysis of the adult Drosophila central brain. eLife 9, e57443 (2020).
Dorkenwald, S. et al. Neuronal wiring diagram of an adult brain. Nature 634, 124–138 (2024).
Hildebrand, D. G. C. et al. Whole-brain serial-section electron microscopy in larval zebrafish. Nature 545, 345–349 (2017).
Turner, M. H., Mann, K. & Clandinin, T. R. The connectome predicts resting-state functional connectivity across the Drosophila brain. Curr. Biol. 31, 2386–2394 (2021).
Randi, F., Sharma, A. K., Dvali, S. & Leifer, A. M. Neural signal propagation atlas of Caenorhabditis elegans. Nature 623, 406–414 (2023).
Shiu, P. K. et al. A Drosophila computational brain model reveals sensorimotor processing. Nature 634, 210–219 (2024).
Pospisil, D. A. et al. The fly connectome reveals a path to the effectome. Nature 634, 201–209 (2024).
Zarin, A. A., Mark, B., Cardona, A., Litwin-Kumar, A. & Doe, C. Q. A multilayer circuit architecture for the generation of distinct locomotor behaviors in Drosophila. eLife 8, e51781 (2019).
Keller, A. J. et al. A disinhibitory circuit for contextual modulation in primary visual cortex. Neuron 108, 1181–1193 (2020).
Kohn, J. R., Portes, J. P., Christenson, M. P., Abbott, L. F. & Behnia, R. Flexible filtering by neural inputs supports motion computation across states and stimuli. Curr. Biol. 31, 5249–5260 (2021).
Lappalainen, J. K. et al. Connectome-constrained networks predict neural activity across the fly visual system. Nature 634, 1132–1140 (2024).
Eckstein, N. et al. Neurotransmitter classification from electron microscopy images at synaptic sites in Drosophila melanogaster. Cell 187, 2574–2594 (2024).
Barnes, C. L., Bonnéry, D. & Cardona, A. Synaptic counts approximate synaptic contact area in Drosophila. PLoS ONE 17, e0266064 (2022).
Kasai, H., Fukuda, M., Watanabe, S., Hayashi-Takagi, A. & Noguchi, J. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 33, 121–129 (2010).
Bargmann, C. I. & Marder, E. From the connectome to brain function. Nat. Methods 10, 483–490 (2013).
Marder, E. Neuromodulation of neuronal circuits: back to the future. Neuron 76, 1–11 (2012).
Gutierrez, G. J., OʼLeary, T. & Marder, E. Multiple mechanisms switch an electrically coupled, synaptically inhibited neuron between competing rhythmic oscillators. Neuron 77, 845–858 (2013).
Stroud, J. P., Porter, M. A., Hennequin, G. & Vogels, T. P. Motor primitives in space and time via targeted gain modulation in cortical networks. Nat. Neurosci. 21, 1774–1783 (2018).
Ferguson, K. A. & Cardin, J. A. Mechanisms underlying gain modulation in the cortex. Nat. Rev. Neurosci. 21, 80–92 (2020).
Connors, B. W. & Gutnick, M. J. Intrinsic firing patterns of diverse neocortical neurons. Trends Neurosci. 13, 99–104 (1990).
Kubota, Y., Hatada, S., Kondo, S., Karube, F. & Kawaguchi, Y. Neocortical inhibitory terminals innervate dendritic spines targeted by thalamocortical afferents. J. Neurosci. 27, 1139–1150 (2007).
Perich, M. G. et al. Inferring brain-wide interactions using data-constrained recurrent neural network models. Preprint at bioRxiv https://doi.org/10.1101/2020.12.18.423348 (2021).
Seung, H. S., Sompolinsky, H. & Tishby, N. Statistical mechanics of learning from examples. Phys. Rev. A 45, 6056 (1992).
Saad, D. & Solla, S. A. Exact solution for on-line learning in multilayer neural networks. Phys. Rev. Lett. 74, 4337 (1995).
Gao, P. et al. A theory of multineuronal dimensionality, dynamics and measurement. Preprint at bioRxiv https://doi.org/10.1101/214262 (2017).
Kim, C. M., Finkelstein, A., Chow, C. C., Svoboda, K. & Darshan, R. Distributing task-related neural activity across a cortical network through task-independent connections. Nat. Commun. 14, 2851 (2023).
Russo, A. A. et al. Motor cortex embeds muscle-like commands in an untangled population response. Neuron 97, 953 (2018).
Beiran, M., Dubreuil, A., Valente, A., Mastrogiuseppe, F. & Ostojic, S. Shaping dynamics with multiple populations in low-rank recurrent networks. Neural Comput. 33, 1572–1615 (2021).
Sompolinsky, H., Crisanti, A. & Sommers, H. J. Chaos in random neural networks. Phys. Rev. Lett. 61, 259 (1988).
Clark, D. G., Abbott, L. F. & Litwin-Kumar, A. Dimension of activity in random neural networks. Phys. Rev. Lett. 131, 118401 (2023).
Hamood, A. W. & Marder, E. Animal-to-animal variability in neuromodulation and circuit function. Cold Spring Harb. Symp. Quant. Biol. 79, 21–28 (2014).
Turner-Evans, D. B. et al. The neuroanatomical ultrastructure and function of a biological ring attractor. Neuron 109, 1582 (2021).
Vishwanathan, A. et al. Predicting modular functions and neural coding of behavior from a synaptic wiring diagram. Nat. Neurosci. 27, 2443–2454 (2024).
Kim, S. S., Rouault, H., Druckmann, S. & Jayaraman, V. Ring attractor dynamics in the Drosophila central brain. Science 356, 849–853 (2017).
Noorman, M., Hulse, B. K., Jayaraman, V., Romani, S., Hermundstad, A. M. Maintaining and updating accurate internal representations of continuous variables with a handful of neurons. Nat. Neurosci. 27, 2207–2217 (2024).
Ben-Yishai, R., Bar-Or, R. L. & Sompolinsky, H. Theory of orientation tuning in visual cortex. Proc. Natl Acad. Sci. USA 92, 3844–3848 (1995).
Prinz, A. A., Bucher, D. & Marder, E. Similar network activity from disparate circuit parameters. Nat. Neurosci. 7, 1345–1352 (2004).
Mastrogiuseppe, F. & Ostojic, S. Linking connectivity, dynamics, and computations in low-rank recurrent neural networks. Neuron 99, 609–623 (2018).
Dubreuil, A., Valente, A., Beiran, M., Mastrogiuseppe, F. & Ostojic, S. The role of population structure in computations through neural dynamics. Nat. Neurosci. 25, 783–794 (2022).
Singer, W. Synchronization of cortical activity and its putative role in information processing and learning. Annu. Rev. Physiol. 55, 349–374 (1993).
Koch, C. & Segev, I. The role of single neurons in information processing. Nat. Neurosci. 3, 1171–1177 (2000).
Goaillard, J.-M. & Marder, E. Ion channel degeneracy, variability, and covariation in neuron and circuit resilience. Annu. Rev. Neurosci. 44, 335–357 (2021).
Seung, H. S. Predicting visual function by interpreting a neuronal wiring diagram. Nature 634, 113–123 (2024).
Li, H., Xu, Z., Taylor, G., Studer, C. & Goldstein, T. Visualizing the loss landscape of neural nets. Adv. Neural Inf. Process. Syst. 31, 6391–6401 (2018).
Fort, S. & Jastrzebski, S. Large scale structure of neural network loss landscapes. Adv. Neural Inf. Process. Syst. 32, 6709–6717 (2019).
Simsek, B. et al. Geometry of the loss landscape in overparameterized neural networks: symmetries and invariances. In Proceedings of the International Conference on Machine Learning 9722–9732 (PMLR, 2021).
Gutenkunst, R. N. et al. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput. Biol. 3, e189 (2007).
Daniels, B. C., Chen, Y. J., Sethna, J. P., Gutenkunst, R. N. & Myers, C. R. Sloppiness, robustness, and evolvability in systems biology. Curr. Opin. Biotechnol. 19, 389–395 (2008).
Fisher, D., Olasagasti, I., Tank, D. W., Aksay, E. R. & Goldman, M. S. A modeling framework for deriving the structural and functional architecture of a short-term memory microcircuit. Neuron 79, 987–1000 (2013).
Naumann, E. A. et al. From whole-brain data to functional circuit models: the zebrafish optomotor response. Cell 167, 947–960 (2016).
Otopalik, A. G. et al. Sloppy morphological tuning in identified neurons of the crustacean stomatogastric ganglion. eLife 6, e22352 (2017).
O’Leary, T., Sutton, A. C. & Marder, E. Computational models in the age of large datasets. Curr. Opin. Neurobiol. 32, 87–94 (2015).
Werbos, P. J. Backpropagation through time: what it does and how to do it. Proc. IEEE 78, 1550–1560 (1990).
Kingma, D. P. & Ba, J. L. Adam: a method for stochastic optimization. In Proceedings of the International Conference on Learning Representations (ICLR, 2015).
Paszke, A. et al. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 32, 8026–8037 (2019).
Van Overschee, D. P. & De Moor, B. Subspace Identification for Linear Systems: Theory–Implementation–Applications (Springer Science and Business Media, 2012).
Beiran, M. & Litwin-Kumar, A. Dataset and code for generating the figures of publication Beiran, M., Litwin-Kumar A., Prediction of neural activity in connectome-constrained recurrent networks. Zenodo https://doi.org/10.5281/zenodo.16618353 (2025).
Acknowledgements
We are grateful to L. F. Abbott for helpful discussions and comments on the paper. M.B. and A.L.-K. were supported by the Kavli Foundation, the Gatsby Charitable Foundation (GAT3708), the Burroughs Wellcome Foundation and National Institutes of Health awards R01EB029858 and RF1DA060772. A.L.-K. was supported by the McKnight Endowment Fund. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
M.B. and A.L.-K. conceived the study. M.B. performed simulations and analyses, with contributions from A.L.-K. M.B. and A.L.-K. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Jakob Macke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Related to Fig. 2.
A Teacher as in Fig. 2. The students are trained on a varying number of recorded neurons M. B Average error in the recorded and unrecorded activity between teacher and students. C Left: Error in the network activity for a given student network in a given trial, when M = 20 neurons are recorded. Right: Error in the task-related readout signal. While the recorded neurons have low error, the unrecorded neurons in the student display large deviations. D Analogous to C, when more neurons are recorded, M = 60. In this case, the activity of unrecorded neurons and the readout are well predicted. E Teacher network from panel A receives a strong external two-dimensional time-varying input, fed to a subset of 100 excitatory neurons. Middle: The dimensionality of the activity, measured by the participation ratio, increases with the input. F Error in unrecorded neuronal activity after training student networks to match the input-driven teacher (color dots), compared to the non-driven teacher (grey dots). Fewer recorded neurons are required to predict activity of unrecorded neurons in this example input-driven network. G Input-driven teacher network with different levels of connectivity sparsity and gain heterogeneity. Teachers have E-I random connectivity, and are initialized at the fixed point. A positive input of unit strength is delivered to 5 excitatory neurons. Recorded neurons correspond to excitatory neurons, while unrecorded neurons can be both excitatory or inhibitory. Teacher networks are generated with different fractions f of non-zero weights, and different ranges for the uniformly distributed gains. Both gains and biases are trained in the students. H Error in unrecorded activity after training vs number of recorded neurons, for different level of sparsity f and gain distributions. While the overall magnitude of the error changes for different gain strengths, the decay of the error as a function of M does not change.
Extended Data Fig. 2 Teacher networks with different dynamics, related to Fig. 3.
A Teachers with variable network size and fixed rank-two connectivity, generating a limit cycle. Right: Error in the activity of recorded neurons after training. The students always learn the dynamics of the teacher. B Error in the single-neuron gains after training. C Example of error in the activity of a recorded neuron and an unrecorded neuron, when there is only one recorded neuron (left), compared to when 7 neurons are recorded (right). For one recorded neuron, the student learns the frequency of the limit cycle, but the temporal profile of the unrecorded neurons does not much the profile of the teacher network. Example for N = 400. D Teachers with variable network size and random connectivity, generating chaotic dynamics. Right: Error in the activity of recorded neurons after training. The students always learn the dynamics of the teacher. E Error in the single-neuron gains for the chaotic teachers. Note that the single-neuron parameters are much better inferred given enough recorded neurons when the teacher is chaotic than when it is low-rank, because there are many more stiff dimensions. F Traces of one example neuron in teacher and student networks with size N = 400 (left) and N = 1000 (right). For N = 400, M = 64 recorded neurons is sufficient to accurately match unrecorded neural activity from the teacher (gray line), while for N = 1000, M = 64 recorded neurons is insufficient but M = 256 is sufficient.
Extended Data Fig. 3 Training connectivity with model mismatch, related to Fig. 4.
A Teacher with model mismatch in the activation function, from Fig. 4a-c. B Example traces of one recorded neuron and one unrecorded neuron in the teacher and after training the student with mismatch in the β parameter. The students networks were trained with 20 recorded neurons (left) and with 150 recorded neurons (right). C Teacher-student framework with mismatch. We train the connectivity of the student, given the teacher’s connectivity as initial condition. The single-neuron parameters are the same in teacher and student, while there is a mismatch in the activation function. Same network as in Fig. 4. D The activation function is a smooth rectification but with different degrees of smoothness, parameterized by a parameter β. Teacher RNN from Fig. 2. E Errors in the activity of recorded (left) and unrecorded (right) neurons for different values of model mismatch between teacher and student. We observe a minor decrease in the error in unrecorded neurons when recording from a large number of neurons, M ≈ 150. F Error in the recorded activity (loss function) for three different mismatch values as a function of training epochs (β = 1. means no mismatch). G Error in the unrecorded activity (loss function) for three different mismatch values as a function of training epochs. H Removing the mismatch in activation by training an additional parameter. We train a student network with the same connectivity as the teacher and different single-neuron parameters. However, the student also does not know the smoothness parameter β. The trained parameters are therefore the gains and biases of each neuron and the smoothness β. I Error in unrecorded activity after training on a subset of M recorded units, similar to C. Training the smoothness parameter of the nonlinearity provides the student with the same prediction power as students without mismatch (see Fig. 2). J Estimated parameter β during training (average and SEM over 10 different initializations). Networks do not retrieve the exact teacher value (β* = 1) although converge to values not far from it on average. Students have a bias towards estimating sharper activation functions (β > 1). Both bias and variance are reduced as the number of recorded neurons is increased.
Extended Data Fig. 4 Dimensionality of the activity and rank of connectivity in the data-constrained RNNs, related to Fig. 5.
A Neural activity traces (centered) used for training the student networks for the three different data constrained RNNs: the premotor network in the Drosophila larva, the central complex in the adult Drosophila, and the oculomotor integrator in larval zebrafish. Different trials/conditions have been concatenated. B Left: First eigenvalues of the covariance spectrum of the datasets. Right: Participation ratio of the activity covariance. The dimensionality of neural activity is higher in the premotor system, then the CX and then the premotor network, indicated by how fast the eigenvalues decay. C Left: Singular values of the connectivity matrix. Right: Estimated rank of the connectivity matrix J, calculated using the participation ratio of the distribution of singular values of J. Given the sparsity and heterogeneity in connectomes, the rank of the connectivity is high.
Supplementary information
Supplementary Information
Supplementary Note
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Beiran, M., Litwin-Kumar, A. Prediction of neural activity in connectome-constrained recurrent networks. Nat Neurosci 28, 2561–2574 (2025). https://doi.org/10.1038/s41593-025-02080-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-025-02080-4