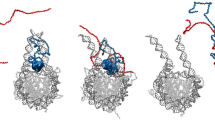
Abstract
Phosphorylation of histone H3 threonine 3 (H3T3) by Haspin recruits the chromosomal passenger complex to the inner centromere and ensures proper cell cycle progression through mitosis. The mechanism by which Haspin binds to nucleosomes to phosphorylate H3T3 is not known. Here we report cryogenic electron microscopy structures of the human Haspin kinase domain bound to a nucleosome. In contrast with previous structures of histone-modifying enzymes, Haspin solely contacts the nucleosomal DNA, inserting into a supergroove formed by apposing major grooves of two DNA gyres. This binding mode provides a plausible mechanism by which Haspin can bind to nucleosomes in a condensed chromatin environment to phosphorylate H3T3. We identify key basic residues in the Haspin kinase domain that are essential for phosphorylation of nucleosomal histone H3 and binding to mitotic chromatin. Our structural data provide notable insight into a histone-modifying enzyme that binds to nucleosomes solely through DNA contacts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Models and cryo-EM maps were deposited in the PDB and Electron Microscopy Data Bank (EMDB) under the following accession codes: Haspin position 1 (PDB: 9B2S, EMDB: 44113), Haspin position 2 (PDB: 9B2T, EMDB: 44114) and Haspin local refinement (PDB: 9B2U, EMDB: 44115). Raw cryo-EM movies were deposited in the EMPIAR database under the accession code EMPIAR-11971. Source data are provided with this paper.
References
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Wang, F. & Higgins, J. M. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol. 23, 175–184 (2013).
Wilkins, B. J. et al. A cascade of histone modifications induces chromatin condensation in mitosis. Science 343, 77–80 (2014).
Yamagishi, Y., Honda, T., Tanno, Y. & Watanabe, Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330, 239–243 (2010).
Broad, A. J., DeLuca, K. F. & DeLuca, J. G. Aurora B kinase is recruited to multiple discrete kinetochore and centromere regions in human cells. J. Cell Biol. 219, e201905144 (2020).
Polioudaki, H. et al. Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett. 560, 39–44 (2004).
Qian, J., Lesage, B., Beullens, M., Van Eynde, A. & Bollen, M. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr. Biol. 21, 766–773 (2011).
Wang, F. et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330, 231–235 (2010).
Kelly, A. E. et al. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330, 235–239 (2010).
Carmena, M., Wheelock, M., Funabiki, H. & Earnshaw, W. C. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13, 789–803 (2012).
Crosio, C. et al. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22, 874–885 (2002).
Welburn, J. P. et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore–microtubule interface. Mol. Cell 38, 383–392 (2010).
Dai, J. & Higgins, J. M. Haspin: a mitotic histone kinase required for metaphase chromosome alignment. Cell Cycle 4, 665–668 (2005).
Dai, J., Sultan, S., Taylor, S. S. & Higgins, J. M. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19, 472–488 (2005).
Higgins, J. M. Haspin‐like proteins: a new family of evolutionarily conserved putative eukaryotic protein kinases. Protein Sci. 10, 1677–1684 (2001).
Higgins, J. Structure, function and evolution of haspin and haspinrelated proteins, a distinctive group of eukaryotic protein kinases. Cell. Mol. Life Sci. 60, 446–462 (2003).
Wang, F. et al. Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. J. Cell Biol. 199, 251–268 (2012).
Dai, J., Sullivan, B. A. & Higgins, J. M. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell 11, 741–750 (2006).
Markaki, Y., Christogianni, A., Politou, A. S. & Georgatos, S. D. Phosphorylation of histone H3 at Thr3 is part of a combinatorial pattern that marks and configures mitotic chromatin. J. Cell Sci. 122, 2809–2819 (2009).
Kawashima, S. A., Yamagishi, Y., Honda, T., Ishiguro, K.-I. & Watanabe, Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327, 172–177 (2009).
Liang, C. et al. Centromere-localized Aurora B kinase is required for the fidelity of chromosome segregation. J. Cell Biol. 219, e201907092 (2020).
Hadders, M. A. et al. Untangling the contribution of Haspin and Bub1 to Aurora B function during mitosis. J. Cell Biol. 219, e201907087 (2020).
Kang, T.-H. et al. Mitotic histone H3 phosphorylation by vaccinia-related kinase 1 in mammalian cells. Mol. Cell. Biol. 27, 8533–8546 (2007).
Jeong, M.-W., Kang, T.-H., Kim, W., Choi, Y. H. & Kim, K.-T. Mitogen-activated protein kinase phosphatase 2 regulates histone H3 phosphorylation via interaction with vaccinia-related kinase 1. Mol. Biol. Cell 24, 373–384 (2013).
Cartwright, T. N. et al. Dissecting the roles of Haspin and VRK1 in histone H3 phosphorylation during mitosis. Sci. Rep. 12, 11210 (2022).
Quadri, R., Sertic, S. & Muzi-Falconi, M. Roles and regulation of Haspin kinase and its impact on carcinogenesis. Cell. Signal. 93, 110303 (2022).
Eswaran, J. et al. Structure and functional characterization of the atypical human kinase haspin. Proc. Natl Acad. Sci. USA 106, 20198–20203 (2009).
De Antoni, A., Maffini, S., Knapp, S., Musacchio, A. & Santaguida, S. A small-molecule inhibitor of Haspin alters the kinetochore functions of Aurora B. J. Cell Biol. 199, 269–284 (2012).
Wotring, L. L. & Townsend, L. B. Study of the cytotoxicity and metabolism of 4-amino-3-carboxamido-1-(β-d-ribofuranosyl) pyrazolo [3, 4-d] pyrimidine using inhibitors of adenosine kinase and adenosine deaminase. Cancer Res. 39, 3018–3023 (1979).
Fedorov, O. et al. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc. Natl Acad. Sci. USA 104, 20523–20528 (2007).
Amoussou, N. G., Bigot, A., Roussakis, C. & Robert, J.-M. H. Haspin: a promising target for the design of inhibitors as potent anticancer drugs. Drug Discov. Today 23, 409–415 (2018).
Tanaka, H. et al. Identification and characterization of a haploid germ cell-specific nuclear protein kinase (Haspin) in spermatid nuclei and its effects on somatic cells. J. Biol. Chem. 274, 17049–17057 (1999).
Tanaka, H. et al. Cloning and characterization of human haspin gene encoding haploid germ cell-specific nuclear protein kinase. Mol. Hum. Reprod. 7, 211–218 (2001).
Villa, F. et al. Crystal structure of the catalytic domain of Haspin, an atypical kinase implicated in chromatin organization. Proc. Natl Acad. Sci. USA 106, 20204–20209 (2009).
Hanks, S. K. & Hunter, T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification 1. FASEB J. 9, 576–596 (1995).
Maiolica, A. et al. Modulation of the chromatin phosphoproteome by the Haspin protein kinase. Mol. Cell. Proteomics 13, 1724–1740 (2014).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Ghenoiu, C., Wheelock, M. S. & Funabiki, H. Autoinhibition and Polo-dependent multisite phosphorylation restrict activity of the histone H3 kinase Haspin to mitosis. Mol. Cell 52, 734–745 (2013).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Zhou, L., Tian, X., Zhu, C., Wang, F. & Higgins, J. M. Polo‐like kinase‐1 triggers histone phosphorylation by Haspin in mitosis. EMBO Rep. 15, 273–281 (2014).
Goto, Y. et al. Pds5 regulates sister-chromatid cohesion and chromosome bi-orientation through a conserved protein interaction module. Curr. Biol. 27, 1005–1012 (2017).
Horn, V. & van Ingen, H. Recognition of Nucleosomes by Chromatin Factors: Lessons from Data-Driven Docking-Based Structures of Nucleosome–Protein Complexes (IntechOpen, 2018).
McGinty, R. K. & Tan, S. Recognition of the nucleosome by chromatin factors and enzymes. Curr. Opin. Struct. Biol. 37, 54–61 (2016).
Kale, S., Goncearenco, A., Markov, Y., Landsman, D. & Panchenko, A. R. Molecular recognition of nucleosomes by binding partners. Curr. Opin. Struct. Biol. 56, 164–170 (2019).
Edayathumangalam, R. S., Weyermann, P., Gottesfeld, J. M., Dervan, P. B. & Luger, K. Molecular recognition of the nucleosomal ‘supergroove. Proc. Natl Acad. Sci. USA 101, 6864–6869 (2004).
Budziszewski, G. R. et al. Multivalent DNA and nucleosome acidic patch interactions specify VRK1 mitotic localization and activity. Nucleic Acids Res. 50, 4355–4371 (2022).
Berg, O. G., Winter, R. B. & von Hippel, P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 20, 6929–6948 (1981).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 42, 243–246 (2024).
Luger, K., Rechsteiner, T. J. & Richmond, T. J. in Methods in Enzymology Vol. 304, 3–19 (Elsevier, 1999).
Lowary, P. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998).
Makde, R. D., England, J. R., Yennawar, H. P. & Tan, S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 (2010).
Dyer, P. N. et al. in Methods in Enzymology Vol. 375, 23–44 (Elsevier, 2003).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Morgan, M. T. et al. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351, 725–728 (2016).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D 66, 486–501 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D 75, 861–877 (2019).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D 74, 531–544 (2018).
Williams, C. J. et al. MolProbity: more and better reference data for improved all‐atom structure validation. Protein Sci. 27, 293–315 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank D. Sousa, D. Ding and K. Cai for their support with cryo-EM sample preparation and data collection at the Beckman Center for Cryo-EM at Johns Hopkins School of Medicine. We thank the Summer Academic Research Experience (SARE) Program at Johns Hopkins for providing support to host S.J.V. for a summer research experience in the Wolberger lab. We thank the Wolberger lab for their insights and discussions on the paper. This work was supported by National Institute of General Medical Sciences grants R35GM130393 (C.W.), R01GM133897 (A.J.H.) and R01GM114119 (A.J.H.) and National Cancer Institute grants F31CA261154 (C.W.H.), F31CA271743 (S.R.) and R01CA266199 (A.J.H.) of the National Institutes of Health, and by a National Science Foundation Graduate Research Fellowship (A.S.E.).
Author information
Authors and Affiliations
Contributions
C.W.H. performed cryo-EM data processing, structural modeling, map interpretation and in vitro experiments to assay Haspin binding and activity on canonical nucleosomes. C.R.G. performed cell-based experiments on Haspin localization. S.R. assayed Haspin phosphorylation of H3T3 on free histone H3 in vitro and on chromatin in cells. X.Z. prepared Haspin kinase domain mutant plasmid constructs. A.S.E. assayed Haspin binding to tailless nucleosomes. S.J.V. froze and clipped cryo-EM grids for data collection. A.J.H. oversaw execution and interpretation of cell-based experiments. C.W. oversaw all aspects of structure determination, biochemistry and data interpretation. C.W.H. and C.W. wrote the paper, with contributions and feedback from all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editors: Sara Osman and Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology and team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cryo-EM data processing workflow for Haspin (465-798) bound to nucleosome.
General processing pipeline to obtain the final cryoEM maps of Haspin Position 1, Haspin Position 2, and Haspin Local Refinement.
Extended Data Fig. 2 Global and local resolution evaluation of both cryo-EM maps.
a, b) Fourier Shell Correlation (FSC) plots using gold-standard 0.143 cutoffs for both cryo-EM maps of Haspin bound to nucleosome, a) position 1, b) position 2. c, d) Local resolution estimation color depictions for both cryo-EM maps corresponding to Haspin bound to nucleosome, c) position 1, d) position 2. e, f) Cut-away slice view through the center of both Haspin cartoon models showing the fit to their respective cryo-EM maps, e) position 1, f) position 2.
Extended Data Fig. 4 Global and local resolution evaluation of Haspin local refinement cryo-EM map.
a) Fourier Shell Correlation (FSC) plot using fold-standard 0.143 cutoffs for the Haspin local refinement cryo-EM map of Haspin bound to nucleosome. b) Local resolution estimation color depictions for the Haspin local refinement cryo-EM map of Haspin bound to nucleosome. c) Cut-away slice view through the center of Haspin showing the fit of the model to the cryo-EM map.
Extended Data Fig. 5 Cryo-EM structure of H3 tail bound to Haspin is similar to crystal structure of H3 peptide bound to Haspin.
A previously reported crystal structure of Haspin (PDB: 4OUC)36 with bound H3 peptide (royal blue) was superimposed over our cryo-EM map of locally-refined Haspin (orange) containing bound H3 tail (light blue). Structures are depicted in cartoon and atom representation and show the similarity in the binding position of H3.
Extended Data Fig. 6 Haspin binding to canonical and tailless nucleosome.
Electrophoretic mobility shift assay (EMSA) showing binding of wild-type Haspin (465-798) to canonical unmodified nucleosome (199 bp) and tailless unmodified nucleosome (199 bp) at the indicated concentrations.
Supplementary information
Source data
Source Data Fig. 5
Unprocessed western blots and gels.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed blots.
Source Data Extended Data Fig. 6
Unprocessed gel.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hicks, C.W., Gliech, C.R., Rahman, S. et al. Haspin kinase binds to a nucleosomal DNA supergroove. Nat Struct Mol Biol 32, 1030–1037 (2025). https://doi.org/10.1038/s41594-025-01502-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01502-y