Abstract
Secondary active membrane transporters harness the energy of ion gradients to concentrate their substrates. Homologous transporters evolved to couple transport to different ions in response to changing environments and needs. The bases of such diversification and, thus, principles of ion coupling are unexplored. Here, using phylogenetics and ancestral protein reconstruction, we investigated sodium-coupled transport in prokaryotic glutamate transporters, a mechanism ubiquitous across life domains and critical to neurotransmitter recycling in humans by excitatory amino acid transporters from the solute carrier 1 family. By inferring ancestral prokaryotic transporter sequences during a change in the ion-coupling mechanism, we found an evolutionary transition from sodium-dependent to independent substrate binding and transport. Structural and functional experiments on ancestral transporters suggest that the transition involved allosteric mutations, rendering sodium binding dispensable without affecting the ion-binding sites. Allosteric tuning of transporters’ energy landscapes might be a widespread route of their functional diversification.
Similar content being viewed by others
Main
Transport of solutes across cell membranes against their concentration gradients is critical for cellular homeostasis. Secondary active membrane transporters catalyze this energetically unfavorable process using the energy of ions flowing down their electrochemical gradients. The ion-binding sites of many transporters are well characterized. However, the three-dimensional (3D) structural properties of transporters necessary to couple substrate and ion movements, and thereby how transporters harvest electrochemical energy, remain elusive.
In the dicarboxylate/amino acid:cation symporter family, the Na+-coupling mechanism persisted through millions of years, conserved between archaea and humans; mammalian excitatory amino acid transporters (EAATs; belonging to the solute carrier 1 (SLC1) family) use Na+ gradients to clear the neurotransmitter glutamate from the synaptic cleft. Many transporter families, including glutamate transporters, show functional diversification using Na+ or H+ gradients (Fig. 1a)1,2. The cell’s environment and needs provide the selective pressure to use specific ions such that when cell membranes are H+-leaky or Na+ is abundant, Na+-coupled transport predominates. In contrast, in environments where Na+ is scarce, such as in fresh water, certain soils and fermented foods, Na+ coupling is no longer beneficial and is eliminated. We used phylogenetic analysis, ancestral protein reconstruction (APR), and structure-function characterization of inferred ancestral proteins to understand how prokaryotic glutamate transporters transitioned away from Na+ coupling. Contrary to our expectations, the transition did not involve promiscuous ion coupling3,4 or a gradual loss of Na+-binding sites. Instead, allosteric changes first made Na+ binding dispensable to function in a distinct evolutionary step, and only then did the transporters lose their Na+-binding residues.
a, Ion-coupling stoichiometries of characterized concentrative transporters in the glutamate transporter family. b, The architecture of Na+-binding sites in GltPh (PDB 6X15). Bound Na1–Na3 are shown as purple spheres. Dotted lines emphasize the coordinating interactions with sidechains, shown as sticks, and main chain carbonyl oxygens, shown as cartoons. c, Sequence alignment of experimentally characterized prokaryotic homologs and inferred ancestral transporters from d. The numbers above the alignment refer to the Na+-binding sites and ‘A’ refers to side chains coordinating the substrate l-aspartate. Signature sequence changes associated with the transition to H+ coupling are shown in red. d, Reduced ML phylogeny of glutamate transporters in Firmicutes. The collapsed ExtNa (purple), ExtInt (green) and ExtH (orange) clades are proportional to the number of the comprising sequences, which is shown within the clade. Numbers at nodes represent corresponding UFBoot2 values. Nodes corresponding to the reconstructed ancestral sequences are highlighted in the corresponding color and labeled AncNa, Anc*, AncInt and AncH. e, Percentage of sequences in each clade containing all Na+-binding site residues ([S/T]92–93, N310, [M/S]311, [D/E]312 and [D/E]405). f, Percentage of sequences in each clade containing signature H+-coupling residues ([A/L/I/V/M/F]92, L311 and N405).
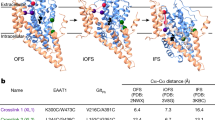
Energetically linked substrate and Na+ binding underlies symport in Na+-coupled glutamate transporter homologs, including archaeal GltPh and GltTk. A higher extracellular Na+ concentration ensures a greater probability of substrate binding from the outside and release on the inside of the membrane, providing directionality to the transport cycle and leading to concentrative uptake5,6,7,8,9,10,11,12. The binding sites for the substrate and three symported Na+ ions are located within a dynamic transport domain, which moves between extracellular and intracellular positions (outward-facing and inward-facing states, Extended Data Fig. 1) in an elevator-like motion relative to a stationary scaffold and trimerization domain13,14,15,16,17,18,19,20,21,22. The transport mechanism and homotrimeric architecture are conserved across glutamate transporters23,24,25,26,27. Na+ binding to two sites (Na1 and Na3) induces the formation of a high-affinity substrate-binding pocket, resulting in substrate and the third Na+ (Na2) binding and closure of the helical hairpin 2 (HP2) gate, which precedes translocation8,9,28,29,30,31,32 (Fig. 1b,c). Thus, ion-coupled binding and transport occur because the high-affinity substrate-binding state is a high-energy state only accessible as Na+ binds. Accordingly, mutations impacting all three Na+-binding sites diminish Na+-coupled substrate binding and transport3,10,33,34,35,36.
Using APR, we reconstructed and characterized ancestral transporter sequences (Fig. 1d); we found that an intermediate ancestral transporter (AncInt) between Na+-coupled and H+-coupled ancestral transporters showed Na+-independent substrate binding and transport, despite preserved Na+-binding sites. Cryo-electron microscopy (cryo-EM) structures showed that AncInt accessed the high-affinity substrate-binding state spontaneously, suggesting that its energy is lower than in Na+-coupled transporters and explaining why Na+ binding was no longer necessary. Detailed evolutionary analysis revealed that the shifted energetic balance from the low-affinity to the high-affinity states was because of allosteric changes distant from the ion-binding and substrate-binding sites, and mutations impacting just two residues in AncInt restored Na+ coupling. We propose that allosterically changing the relative energy of functional states in transporters constitutes an evolutionary mechanism allowing the diversification of strictly specific ion coupling observed in present-day transporters. Our work reveals key principles underlying Na+ coupling, showcases the feasibility and power of structure–function APR in untangling the complex mechanisms of membrane transporters, and suggests a general evolutionary mechanism underlying adaptations to different ionic environments in secondary active transporters.
Results
An evolutionary intermediate between Na+-coupled and H+-coupled transporters
We used phylogenetic analysis and APR to understand how bacterial glutamate transporters might have changed their ion-coupling preference (Extended Data Fig. 2). The approach can reveal minimal subsets of stepwise, chronologically ordered changes required to alter functions, including epistatic changes, which are insufficient for the functional switch but are prerequisites for it37. Subsequent inference of ancestral sequences along an evolutionary pathway provides a platform to test evolutionary and mechanistic hypotheses38,39,40,41,42,43,44. We selected sequences in the Firmicutes/Bacillota phylum to recapitulate the transition from Na+ to H+ coupling, rooting the phylogenetic tree to archaeal Na+-coupled GltPh (Methods and Extended Data Fig. 3a–d). We identified three major monophyletic clades in our tree (Fig. 1d), with two predictably displaying conserved signature sequences of Na+ and H+ coupling (Fig. 1e,f and Extended Data Fig. 3d). We refer to these present-day extant sequences as ExtNa and ExtH clades. Strikingly, we also observed an intermediate clade ExtInt between ExtNa and ExtH clades. Only 16% of its sequences contain all Na+-coordinating residues, and none contain all H+-coupling signature residues (Fig. 1e,f and Extended Data Fig. 3d). This branch was robust to perturbation and reconstructed with high confidence on the basis of a UFBoot2 bootstrap value > 95% (Fig. 1d)45. The extant sequences closest to ExtInt with the full complement of Na+-coordinating residues are referred to as Ext*.
Next, we inferred ancestral sequences by determining posterior probabilities (PPs) of each amino acid occurring at each position (site) in the multiple-sequence alignment (Extended Data Fig. 4a–f). Each node’s final maximum likelihood (ML) amino acid sequence is constructed using the most probable amino acid at every site. We reconstructed sequences corresponding to the closest ancestor to the Na+-coupled root (AncNa) and the ancestor of the intermediate ExtInt clade (AncInt); the average PPs per site were 0.87 and 0.85, respectively (Extended Data Fig. 4a,b), which is sufficiently high for APR studies46,47. As expected, reconstructed AncNa sequences showed signature sequences of Na+-coupled transporters (Fig. 1c and Extended Data Figs. 4a and 5). Surprisingly, AncInt also featured the full complement of Na+-binding residues with high PPs, contrasting with their poor conservation among the descendent present-day transporters (Figs. 1c and 2a and Extended Data Figs. 4a and 5). These observations suggest a lack of selective pressure on Na+-binding residues in AncInt, raising the question of its functional properties.
a, Comparison of the signature Na+-coupling residues in present-day ExtInt transporters and inferred AncInt. ExtInt WebLogo represents the conservation of transporter residues in the ExtInt clade. AncInt WebLogo is a visual representation of the PP per site in the ancestral reconstruction; the topmost residue at each site represents the ML residue. b, Representative SEC profiles of GFP-tagged AncInt purified in ligand-free conditions. Black and dotted green lines correspond to the relative optical absorption at 280 nm and GFP fluorescence (excitation, 485 nm; emission, 510 nm), respectively. Inset, SDS–PAGE analysis of GFP-tagged AncInt, stained with total protein Coomassie stain (total) or imaged by in-gel GFP fluorescence (GFP). ‘Affinity’ refers to elution following streptactin affinity purification. ‘SEC’ refers to peak fractions eluting at 14.5 ml following SEC. c, [3H]l-aspartate uptake in the presence (filled squares) or absence (filled circles) of a Na+ gradient. Left, simplified proteoliposome reaction setup. All experiments were performed at an estimated potential of −102 mV, generated by valinomycin-mediated K+ efflux. Lines represent AncNa (solid purple) or AncInt (dashed green). Open circles represent AncInt without a Na+ gradient and in the presence of 1 mM dl-TBOA. Statistical significance at 10 min was measured using unpaired two-tailed t-tests. ****P < 0.0001 and *P = 0.0193; not significant (NS), P = 0.3431. Error bars are the means ± s.d. of three (AncNa) or four (AncInt) independent experiments, each the average of at least two technical replicates; error bars not shown are smaller than the displayed symbol. d–g, Exchange activity of ancestral transporters. All experimental setups had external 0.5 µM [3H]l-aspartate and internal 500 µM unlabeled l-aspartate; negative controls (clear circles) contained an additional external 500 µM unlabeled l-aspartate. Error bars are the means ± s.d. of three independent experiments, each the average of at least two technical replicates. d,e, Sodium-dependent counterflow of AncNa (d; purple) or AncInt (e; green) in the presence of internal and external 10 mM NaCl (filled squares) or choline chloride (filled circles). f,g, pH-dependent counterflow of GltTBc (f; orange) or AncInt (g; green) in the presence of internal and external KH2PO4 and K2HPO4 at pH 6 (filled squares), 7 (filled circles) or 8 (filled triangles).
To design ancestral constructs for protein expression, we replaced poorly aligned highly variable loops and tails, excluded from the APR analysis, with those from an Ext* sequence with the highest identity to AncInt (Extended Data Fig. 5). To minimize the impact of these regions on our comparisons between ancestors, we used the same loops and tails for all ancestral constructs. Most of these were single-residue insertions, except for the C-terminus and the loop between transmembrane helix 3 (TM3) and TM4. Although these insertions might affect protein stability, flexibility and function, previous studies in GltPh suggest that these regions are not directly involved in ion coupling. The C-terminus has been deleted in structural and functional studies13, and cleavage of the 3–4 loop affected transport kinetics but not ion coupling48. We expressed ancestors in Escherichia coli with or without a C-terminal optimized GFP49 and purified them (Fig. 2b and Supplementary Fig. 1a–c). While there are several bands above and below the primary band, they correlate with GFP fluorescence in gels and size-exclusion profiles. Therefore, these are likely misfolded or aggregated species. Despite extensive optimization, the ancestral proteins expressed ~10-fold lower than extant prokaryotic homologs, yielding ~20–100 µg of purified proteins per L of culture.
The ancestral intermediate transporter is uncoupled from Na+
When we purified and reconstituted AncNa into proteoliposomes, we observed a time-dependent accumulation of [3H]l-aspartate in vesicles in the presence of an inward Na+-gradient. In contrast, no [3H]l-aspartate was retained in the absence of Na+ (Fig. 2c). These results suggest that AncNa is adequately folded and recapitulates expected Na+ coupling. When we repeated the experiment with AncInt, we observed lower [3H]l-aspartate retention unaffected by the removal of Na+ but reduced in the presence of the competitive inhibitor dl-TBOA (Fig. 2c), suggesting that AncInt in proteoliposomes might be able to bind and transport l-aspartate independently of Na+. To demonstrate this, we measured [3H]l-aspartate accumulation in AncInt proteoliposomes under exchange conditions1,50, with 500 µM unlabeled l-aspartate inside the vesicles and 0.5 µM [3H]l-aspartate outside. AncInt accumulated substrate independently of Na+ or pH, unlike the respective controls, AncNa and GltTBc (Fig. 2d–g). Together, this suggests that AncInt functions as a cation-independent exchanger.
We used microscale thermophoresis (MST) to measure substrate-binding affinity to AncInt. AncInt binds l-aspartate with similar micromolar affinities in the absence and presence of 100 mM Na+ (Fig. 3a). Notably, the MST signal-to-noise values upon l-aspartate binding ranged from 8.6 to 19.5, reflecting the high confidence of the binding isotherms (Supplementary Fig. 2a). We confirmed this finding using nano-differential scanning fluorimetry (nanoDSF) to measure ligand-dependent protein melting temperatures (Tm) as a qualitative proxy for ligand binding51,52. Adding l-aspartate to AncInt in the absence of Na+ increased Tm by 7.1 °C, suggesting that, as expected, l-aspartate can bind without Na+ (Fig. 3b,c and Supplementary Fig. 2c). Notably, 10 mM Na+ also stabilized the protein by 5.3 °C without l-aspartate and by 13.5 °C in the presence of 1 mM l-aspartate, suggesting that the ions can still bind to the transporter regardless of substrate binding (Fig. 3c,d and Supplementary Fig. 2c). In AncNa, the MST signal-to-noise ratio was insufficient to accurately determine binding affinities, likely because of changes in measurable parameters, including charge, hydration shell or conformation53. In nanoDSF l-aspartate titrations, much lower concentrations of l-aspartate were required to observe substrate binding in 10 mM Na+ as compared to 0 mM Na+ (Fig. 3e). We observed insignificant or small Tm increases when adding 1 mM l-aspartate or 10 mM Na+ separately compared to the ligand-free AncNa (Fig. 3f,h and Supplementary Fig. 2b). In contrast, we observed a dramatic 11.3 °C increase when adding 1 mM l-aspartate in the presence of 10 mM Na+(Fig. 3f,g). Thus, l-aspartate and Na+ bind to AncNa cooperatively, as reported for Na+-coupled transporters1,8,10,11,12,54,55.
a, l-Aspartate binding by AncInt in 100 mM NaCl (green circles) or 100 mM choline chloride (black circles), measured by MST. The data shown are the means ± s.d. from three independent experiments; however, the first two points (squares) had points excluded because of aggregation and, therefore, do not have error bars. The best-fit KD values and s.e.m. in NaCl and choline chloride were 27 ± 3 µM and 35 ± 5 µM, respectively. b, AncInt Tm in buffer alone (ligand-free), 1 mM l-aspartate, 10 mM NaCl or both NaCl and l-aspartate. The data shown are from two independent experiments, each with several technical replicates. c,d, AncInt ΔTm values upon addition of l-aspartate in the presence and absence of Na+ (c) and in the presence of Na+ but absence of l-aspartate (d). e, Tm of AncNa as a function of l-aspartate concentration in either 0 mM NaCl (black circles) or 10 mM NaCl (purple circles). Lines are to guide the eye and represent the means of the two displayed independent replicates. f, AncNa Tm in buffer alone (ligand-free), 1 mM l-aspartate, 10 mM NaCl or both NaCl and l-aspartate. The data shown are from two independent experiments, each with several technical replicates. g,h, AncNa ΔTm values upon the addition of l-aspartate in the presence and absence of Na+ (g) and in the presence of Na+ but absence of l-aspartate (h); data are from two independent experiments.
These results suggest that, unlike in Na+-coupled transporters, l-aspartate and Na+ can bind to AncInt independently; therefore, substrate and ion binding are ‘uncoupled’. Notably, while we did not observe any Na+ stimulation in either MST or exchange experiments, the ΔTm of l-aspartate was larger in the presence of Na+ (15.3 °C versus 7.1 °C), suggesting residual cooperativity. However, thermal melts are not thermodynamic measurements and ΔTm values cannot be directly related to ligand affinity. Overall, we conclude that the intermediate ExtInt clade functionally diverged from the Na+-coupled ExtNa transporters by uncoupling substrate from Na+ binding. We attempted to verify that the reconstructed AncInt was robust to phylogenetic uncertainty using ‘AltALL’ sequences where all ambiguous residues are varied46 but these produced misfolded proteins. Instead, we validated the observed functional divergence by examining the necessary and sufficient sequence changes, as discussed below.
AncInt accesses high-affinity states without Na+
To visualize how Na+-independent aspartate binding occurs, we purified AncInt, reconstituted it into MSP1E3 lipid nanodiscs (Supplementary Fig. 3), exchanged it into a buffer containing no Na+ or any other alkali cations (20 mM HEPES pH 7.4 and 100 mM NMDG-Cl), and froze cryo-EM grids after and before adding 1 mM l-aspartate (that is, in bound and ligand-free conditions, respectively) (Supplementary Figs. 4 and 5). As a Na+-coupled control, we reconstituted GltPh into MSP1E3 lipid nanodiscs and froze grids under ligand-free conditions (Supplementary Fig. 6).
First, we compared the structures of GltPh and AncInt in ligand-free conditions. Cryo-EM imaging of GltPh revealed the presence of two structural classes with final maps at 2.7-Å and 3.1-Å resolution (Extended Data Figs. 6 and 7, Supplementary Fig. 6 and Table 1). A highly conserved nonhelical loop N310-M311-D312 (NMD) motif, which breaks TM7 into TM7a and TM7b, is the nexus for solute binding and participates in coordinating Na1, Na2, Na3 and substrate through its side chain and main chain atoms. In both classes, the NMD motif is in a ‘flipped-out’ configuration, which distorts substrate-binding and Na+-binding sites (Fig. 4a,e). We call these conformations, characteristic of ligand-free Na+-coupled transporters, ‘low-affinity’ states9,15,16,30,56. The two ligand-free GltPh structural classes differ in the position of the transport domain. One corresponds to the outward-facing state (Fig. 4a,e) and the other corresponds to a previously observed intermediate16,21,30,57 where the transport domain moves toward the inward-facing state by about one quarter of the way (Extended Data Figs. 7 and 8a). The HP2 gate is open in the outward-facing states, propped open by M311, and closed in in the intermediate, with M311 pointing out into the lipid from underneath (Fig. 4a and Extended Data Fig. 8a).
All numbered residues use GltPh numbering. All structures are aligned on the HP1/TM7a region (residues 259–309 in GltPh). All densities are in gray and are sharpened and contoured at 10σ. Purple spheres represent bound Na+ ions, and dashed circles represent unoccupied Na+ sites. Na+-coupled substrate binding of GltPh (purple; a–g) and uncoupled substrate binding of AncInt (green; h–n). Panels depict the substrate-binding (a–d,h–k) and ion-binding (e–g,l–n) sites. a,e, In ligand-free GltPh (outward-facing), both substrate-binding and ion-binding sites are distorted, forming a low-affinity configuration. b,f, Superposition of Na+-bound GltPh (PDB 7AHK) and translucent ligand-free GltPh. Upon Na1 and Na3 binding, the NMD loop and M311 side chain flip into the pocket. TM3 (S92-T93) moves closer to the NMD loop to form the Na3 and Na1 sites and the M311, R397 and N401 side chains adjust to form Na2-binding and l-aspartate-binding sites. These changes produce the high-affinity substrate-binding state. c,d,g, Na+/l-aspartate-bound GltPh (PDB 6X15) demonstrates subsequent l-aspartate/Na2 binding and HP2 closure; l-aspartate coordination was determined by hbonds in ChimeraX. h,i,l,m, In ligand-free conditions, AncInt visits both low-affinity (h,l) and high-affinity (i,m) states. Spontaneous NMD loop and side-chain movements are similar to those induced by Na+ binding in GltPh (translucent purple, l,m). j,k,n, l-Aspartate binding to the preformed site and HP2 closure in AncInt without Na+; l-aspartate coordination is similar to GltPh.
Previously determined GltPh and GltTk structures show that, upon Na+ binding to Na1 and Na3, Na+-coupled transporters restructure from the low-affinity ligand-free state to a ‘high-affinity’ state through allosterically coupled changes8,9,28,10 (Fig. 4b,f). Their NMD motifs go from the flipped-out to the flipped-in configuration, repositioning M311 proximal to the substrate (Fig. 4b) and altering the tilt of TM7b30. R397 moves from occupying the substrate-binding site to forming a cation–π interaction with Y317 in TM7b (Fig. 4b), with its guanidium group in place to salt-bridge with l-aspartate. Moreover, N401 repositions slightly to coordinate l-aspartate optimally30. Together, these changes underlie high-affinity binding by optimally positioning residues coordinating the substrate and the additional Na2 (Fig. 4c,d,g). Following substrate binding, HP2 closes (Fig. 4g), facilitating translocation of the transport domain to the inward-facing state.
Cryo-EM imaging of ligand-free AncInt revealed three outward-facing structural classes, all refined to 3.0-Å resolution (Extended Data Fig. 7, Supplementary Fig. 5 and Table 1). One class resembles the low-affinity ligand-free GltPh with a flipped-out NMD motif and an empty binding pocket; however, R397369 is already in place to coordinate l-aspartate (hereafter, regular text and subscript correspond to GltPh and AncInt numbering, respectively) (Fig. 4h). The Na+-binding sites are distorted similarly to ligand-free Na+-coupled transporters (Fig. 4l), except that T9278 and [T/S]9379 in the Na3 site are already correctly positioned.
However, unlike ligand-free GltPh, we also observed two ligand-free AncInt classes in high-affinity states, with substrate-binding and Na+-binding site architectures like those of GltPh after Na+ binding to Na1 and Na3 sites (Fig. 4i,m). The NMD motifs are flipped in, R397369 and N401373 are correctly oriented for l-aspartate coordination (Fig. 4i), and all residues in Na1 and Na3 sites are configured like in Na+-bound GltPh (Fig. 4m). Water molecules likely occupy the sites in the absence of Na+ but the resolution is insufficient to model them, and computational studies will be needed to understand how they compensate for the missing ions. One of the classes shows an HP2 structurally identical to substrate-bound AncInt, except with an unresolved tip (the loop connecting the two helical arms of the hairpin) and an empty substrate-binding pocket (Extended Data Fig. 8d). In the other class, an ambiguous density occupies the pocket that is too large for l-aspartate and might correspond to a spuriously bound buffer component; this site might be generally attractive to anions28 (Extended Data Fig. 8b). We excluded this structure and inward-facing classes from further consideration; the latter are poorly resolved and have notable regions of missing density in the transport domain (Extended Data Fig. 7). We expect similar structures of the substrate-binding and ion-binding sites in the inward-facing and outward-facing states on the basis of observations in other glutamate transporter family members15,16,30.
Binding sites are unaltered in uncoupled transporters
Imaging of aspartate-bound AncInt in Na+-free conditions revealed a single outward-facing structural class, which we refined to 3.4-Å overall resolution (Extended Data Figs. 6 and 7, Supplementary Fig. 4 and Table 1). The only structural differences between aspartate-bound AncInt and high-affinity ligand-free AncInt are that its HP2 tip is well structured and closed, and the substrate-binding pocket is occupied by a strong nonprotein density, which we modeled as l-aspartate (Fig. 4j,k,n and Extended Data Fig. 8e). The substrate orientation and binding site architecture of AncInt in the absence of alkali cations were nearly identical to Na+/aspartate-bound GltPh (Fig. 4d,k). The NMD motif is flipped in with the M311289 side chain pointing into the substrate-binding pocket and forming van der Waals interactions with l-aspartate, which is coordinated by side chains of conserved T314292, D394366, R397369 and N401373 (Fig. 4k). R397369 is the principal residue interacting with the side-chain carboxylate of l-aspartate and D394366 coordinates the amino group; these residues determine the specificity of the transporter for the acidic over neutral amino acids and dicarboxylates, respectively58,59,60. N401373, one of the most conserved residues throughout the evolution of glutamate transporters, coordinates the main chain carboxylate of l-aspartate. At the current resolution, the architectures of all three Na+-binding sites in AncInt are identical to Na+/aspartate-bound GltPh (Fig. 4n), although Na+ is absent.
Our results suggest that ligand-free AncInt spontaneously populates the high-affinity state with the correctly formed substrate-binding and ion-binding sites, facilitating substrate binding without Na+ binding. In contrast, this is a high-energy state in GltPh and other Na+-coupled transporters, only accessible upon Na+ binding.
Allosteric mutations turn ion coupling back on
The amino acid sequence changes underlying the transition from Na+ coupling in AncNa to independence in AncInt are allosteric and lie outside the ion-binding sites. AncNa and AncInt are 78% identical over the transport domain sequence (Extended Data Fig. 4c), with a difference in 26 amino acids and PPs above 0.9 in at least one of the ML sequences (Extended Data Fig. 4d). The changes, however, are subtle, making it difficult to pinpoint those responsible for gaining independence from Na+ ions. Our phylogenetic tree shows that the evolution from Na+-coupled to uncoupled function occurred through several ancestral intermediates between AncNa and AncInt, which might have gradually acquired the mutations. To narrow down the search for changes necessary for the functional switch, we examined the ancestor immediately preceding AncInt, Anc*, and a present-day transporter most homologous to Anc*, Ext* (KJS87745.1) (Fig. 1d). Anc* and Ext* showed robust Na+-coupled binding and uptake, respectively, suggesting they are Na+ coupled (Fig. 5a and Extended Data Fig. 9).
a, [3H]l-aspartate uptake of Ext* in the presence (filled squares) or absence (filled circles) of a Na+ gradient, with the same reaction setup as in Fig. 2c. Error bars are the means ± s.d. of three independent experiments, each the average of at least two technical replicates; error bars not shown are smaller than the displayed symbol. b, Transport domain of l-aspartate-bound AncInt, with unoccupied Na+-binding sites shown as light-gray spheres, TM7 in blue and P295273 and Q327305 as black spheres. c, [3H]l-Aspartate uptake of AncInt P295273S and Q327305N with the same reaction setup as Figs. 2c and 4a. Error bars are the means ± s.d. of three independent experiments, each the average of at least two technical replicates. In each individual experiment, the first time point in choline chloride conditions (15 s) was set to zero. d, Tm of AncInt P295273S and Q327305N in buffer alone (ligand-free), 10 mM NaCl, 1 mM l-aspartate or both NaCl and l-aspartate. Replicates are from two independent experiments, each with several technical replicates. e, Comparison between the low-affinity and high-affinity GltPh (e–h) or AncInt (i–l) structures aligned on the rigid HP1 (GltPh residues 258–292). Darker structures are the high-affinity states (Na+/aspartate-bound GltPh (PDB 7RCP) and aspartate-bound AncInt), with the NMD loop positioned toward the substrate-binding site. Lighter structures are ligand-free GltPh and low-affinity ligand-free AncInt, with the NMD loop positioned away from the substrate-binding site. All residue numbering corresponds to GltPh numbering. e,i, TM7 alone with P295273 and Q327305 shown as white and black spheres in low-affinity (e) and high-affinity (i) states; view corresponds to the rotated (right) view in Fig. 4b. f,j, A close-up view of the NMD loop (f) and the start of TM7b (j). Note the larger shift of TM7b in GltPh than AncInt. g,k, a view of the transport domain from the extracellular space showing larger packing differences between low-affinity (g) and high-affinity (k) conformations in GltPh. h,l, Close-up views of [N/Q]327305 in TM7 interacting with the top of TM3 in AncInt.
There are differences in 18 amino acids between Anc* and AncInt (95.6% identical), and only two of these are located in the transport domain and have high PPs: S295273P (0.969) and N327305Q (0.996). These residues are positioned at the beginning and end of TM7, respectively (Fig. 5b). We hypothesized that the reverse substitutions in AncInt, P295273S and Q327305N, would be sufficient to switch ion coupling back on. Indeed, in proteoliposomes, the mutant exhibited Na+-dependent concentrative l-aspartate transport (Fig. 5c). In the thermal shift assays, l-aspartate stabilized the mutant when added with Na+ ions but not without them (Fig. 5d and Supplementary Fig. 7). Na+ alone induced a deeper and sharper first-derivative trough, consistent with ions binding and increasing the cooperativity of the unfolding process. Both substitutions appear to be necessary, as single mutants showed changes in the melting curves suggesting that the substrate binds without Na+ (Extended Data Fig. 9).
We also collected a cryo-EM dataset on AncInt P295273S;Q327305N in ligand-free conditions. At the achieved resolution of 5.0 Å, we could not model side chains and the NMD loop; nevertheless, the density of all observed structural classes aligned well with the TM3, TM7b, HP2 and TM8a regions of the low-affinity but not the high-affinity structures of AncInt (Supplementary Fig. 8). Together, these results suggest that substitutions can rescue Na+ coupling in AncInt by shifting the equilibrium of the ligand-free transporter back to the low-affinity state, thereby restoring coupled substrate and Na+ binding. Interestingly, introducing the forward substitutions (S295273P and N327305Q) into AncNa did not turn off Na+ coupling (Extended Data Fig. 9). Furthermore, introducing these substitutions in Anc* and Ext* resulted in a misfolded, unstable protein. Thus, TM7 mutations are necessary but insufficient to turn ion coupling off. Additional mutations might be required to compensate for negative fitness effects and shift energetics to complete uncoupling.
We compared the low-affinity and high-affinity states of Na+-coupled GltPh (Fig. 5e–h) and uncoupled AncInt (Fig. 5i–l) to understand how allosteric mutations might affect their equilibrium. TM7 runs through the core of the transport domain with the NMD motif at its center (Fig. 5b,e,i). In Na+-coupled transporters GltPh and human EAAT3, TM7b shows a displacement in the low-affinity ligand-free state compared to the high-affinity Na+-bound state (Fig. 5e,f and Supplementary Table 1), translating into repacking of the surrounding TM3, TM6, TM8 and HP2 (Fig. 5g,h). Packing differences in this region affect GltPh affinity for l-aspartate and are, therefore, allosterically coupled to the changes at the binding site3,61,62. In contrast, TM7b of AncInt shows minimal tilt changes and less helical repacking in the low-affinity state compared to the high-affinity state (Fig. 5i–k), suggesting that S295273P and N327305Q substitutions stabilize the high-affinity state. P295273 is in the loop preceding TM7a and might rigidify the loop compared to serine (Fig. 5b,i). Q327305 is the last residue of a highly conserved F-[I/V]-A-[N/Q] signature sequence motif in TM7b63 (Fig. 5b,i). Q327305 hydrogen-bonds to the top of TM3 (Fig. 5l), perhaps stabilizing TM7–TM3 interactions. By contrast, N327305 in GltPh is too short to reach TM3 (Fig. 5h). These analyses suggest that the structural flexibility of the central TM7 determines the equilibrium between the transporter states, and a more rigid TM7 in AncInt tightly packed to surrounding helices might eliminate the need for Na+ binding to achieve the high-affinity state.
Discussion
Na+-coupled glutamate transport is one of the most thoroughly characterized ion-coupled transport mechanisms, where Na+ binding is inextricably linked to substrate binding. Yet, how these binding sites are allosterically coupled, and thus the design principles of Na+-coupled transporters, were unknown. We gained unprecedented insights into this fundamental question by examining the evolution of prokaryotic glutamate transporters using phylogenetics and APR. Surprisingly, the transition from powering transport with Na+-to-H+ gradients occurred through an intermediate ancestral transporter AncInt, in which l-aspartate binding and transport were uncoupled from Na+ binding. Our reconstructed ancestors seem to show slow transport compared to extant transporters, although we did not systematically study this. Reconstructed ancestral proteins are generally more thermostable than extant proteins, which is an established artifact of APR4,64. This thermostability might manifest as slow transitional kinetics and slow transport rates.
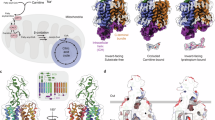
Unlike Na+-coupled homologs, AncInt does not require Na+ to form high-affinity substrate-binding and ion-binding sites. Thus, we propose that the relative free energies of the low-affinity and high-affinity states are the linchpin of ion coupling (Fig. 6a–c). In Na+-coupled transporters, the free energy of the low-affinity conformation is much lower than the high-affinity conformation (Fig. 6b). The substrate-binding energy alone is insufficient to overcome the penalty, and Na+ binding pays the energetic price, stabilizing the state. In uncoupled AncInt, the low-affinity state has a similar or higher energy, and the high-affinity state is populated without ions (Fig. 6c). Na+ ions can still bind, possibly with higher affinity because of preformed Na+ sites. However, they are unnecessary for substrate binding.
a, The low-affinity (yellow square) and high-affinity (blue circle) binding configurations highlight changes in R397, N401 and the NMD loop conformations within the binding site. The square and circle represent the differences in the transport domains outside the binding sites allosterically coupled to the conformations of R397, N401 and the NMD loop. The dotted ovals represent local configurations that are unsuitable (red) or ready (green) to coordinate l-aspartate (green solid oval). b,c, The hypothetical transporter energy landscapes modulated by Na+ binding of Na+-coupled (b) and uncoupled (c) transporters. Solid lines represent the relative energies of the low-affinity and high-affinity states in the absence of Na+; the dashed line represents the changes upon Na+ binding. Relative to the high-affinity state (blue circle), the energy of the low-affinity state (yellow square) in Na+-coupled transporters is lower than in uncoupled transporters because of allosteric mutations outside the binding sites. In Na+-coupled transporters, Na+ binding lowers the energy of the high-affinity state, promoting substrate binding. In Na+-uncoupled transporters, Na+ binding is unnecessary. This image was created with BioRender.com.
We found two high-probability allosteric mutations between uncoupled AncInt and the previous ancestor inferred to be Na+ coupled. In AncInt, these were sufficient to restore Na+-coupled binding and transport, likely by stabilizing the low-affinity state. However, it seems likely that before this switch, transporters accumulated other neutral epistatic mutations during the evolution from Na+-coupled AncNa to AncInt, which gradually increased the relative energy of the low-affinity state. Then, the final two changes were sufficient to make the high-affinity state accessible and function independent of Na+ (Fig. 6c). Such allosteric alterations of energy landscapes might underlie the evolutionary diversification of many proteins65,66,67,68. This mechanism explains how a few mutations can turn off Na+ coupling without requiring several mutations to eliminate the three Na+-binding sites, which would diminish the transporter’s ability to bind substrate and overall cellular fitness.
Was the transition to uncoupled substrate binding a random evolutionary event or an adaptation required for organismal fitness? While functional studies of AncInt descendants will need to be carried out, most are unlikely to be ion coupled according to the lack of Na+-coupled or H+-coupled signature residues. Transporter copies or transporters from different families with similar functions could compensate for the loss of ion-coupled transport69, which still raises the question of the physiological relevance of uncoupled transporters. Examining the gene neighbors of uncoupled transporters revealed that many are adjacent to enzymes involved in amino acid metabolism; we observed no similar preferences in the Na+-coupled or H+-coupled gene neighbors (Extended Data Figure 10). Thus, uncoupled transporters might mediate amino acid uptake paired with their enzymatic use, ultimately maintaining the inward concentration gradient required for continuous uncoupled transport. Such pairing would be reminiscent of glucose transporters, which pair uncoupled transport with subsequent glucose metabolism, a key interplay for insulin secretion70.
In summary, we show that phylogenetic analysis and APR can uncover the evolutionary trajectory of functional diversification in transporters and pinpoint amino acid changes that alter their function. Comparative studies of ion-coupled and ion-independent proteins show that ion coupling can be allosterically regulated and turned on and off by mutations and perhaps environmental changes, which reshape the energy landscape of the transporter. We suggest that drugs could also allosterically modulate the extent of ion and substrate coupling in secondary active transporters, allowing a novel inhibitory modality in which transport is not abolished but its concentrative capacity is diminished. Such drugs could offer advantages in cases where complete inhibition might be harmful.
Methods
Sequence retrieval, curation and alignment
We designed a workflow to construct ML phylogenies capturing the Na+-coupled to H+-coupled (ExtNa to ExtH) evolutionary transition on the basis of previous APR studies47 (Extended Data Fig. 2). Biochemically characterized ExtH proteins feature signature sequence substitutions in the Na+-binding sites compared to ExtNa transporters2,25,50: [T/S]92[A/L/I/V/M/F], D405N and M311L (numbering is based on ExtNa archaeal transporter GltPh) (Fig. 1c). Putative ExtH genes carrying these signature residues are widespread throughout bacterial phyla. We constructed a maximum-parsimony tree of all bacterial glutamate transporter homologs and found that putative ExtH transporters cluster together regardless of phylum (Extended Data Fig. 3a). Although we cannot exclude the existence of H+-coupled transporters with completely different signature residues, we infer that the emergence of ExtH was a single-gene duplication event, after which ExtH genes spread through horizontal gene transfer.
Aiming to have a sequence set with a size amenable to APR, we retrieved 20,000 nonredundant Bacillota/Firmicutes sequences using PSI-BLAST (National Center for Biotechnology Information) because ExtH sequences were predominantly found in this phylum. We used WP_011876993.1 as the original query, which we estimated would be close to both Na+-coupled and H+-coupled transporters71,72. After clustering by 99% using CD-HIT73, removal of sequences with ambiguous amino acid assignment and alignment by MUSCLE v5 (ref. 74), a maximum-parsimony tree was generated by MP-boot75 with the remaining 9,891 sequences. Subsets of sequences were selected on the basis of proximity to sequences with signatures of H+ coupling and nearby sequences up until those with signature ExtNa residues (Extended Data Fig. 3b), resulting in 5,249 sequences. The remaining sequences were clustered to 90% using CD-HIT and the resulting 2,539 sequences were manually curated to exclude sequences with major gaps or insertions. Archaeal GltPh (UniProt O59010) was included to be the tree root. The final sequence set (1,410 sequences) was aligned by MAFFT-DASH, which incorporates information from Protein Data Bank (PDB) structures into the alignment matrix76. Finally, variable loops and tails were removed and the resulting alignment was manually adjusted to account for poorly aligned regions.
Phylogenetic analysis, ancestral protein reconstruction and construct design
ML trees and ancestral sequences were generated using IQ-TREE 2.1.4-beta (ref. 77). Evaluation of all potential evolution models and rate heterogeneity parameters using ModelFinder revealed that LG + F + R was the best protein evolution model78,79,80. FreeRate (F) heterogeneity considers site-specific differences in evolutionary rate, relaxing the constraint of equal site probabilities imposed by discrete gamma rate heterogeneity. The number of possible rate categories that could be assigned to a site (R) was empirically determined for each run using ModelFinder to query a range of 10–20.
As with other APR studies of microbial proteins, our tree topology is not reconciled to the microbial species tree, as this has poor confidence especially at lower taxonomic ranks81. We chose the outgroup-based approach of rooting the tree, using archaeal GltPh as the outgroup because of the evolutionary distance between bacteria and archaea; GltPh was also distant enough to the ingroups and did not introduce any long-branch attraction artifacts. Most archaeal transporters contain all Na+-coupling signature residues (Extended Data Fig. 3c), suggesting that the origin of glutamate transport was sodium coupled and the divergence into sodium-independent exchangers and subsequent proton-coupled transporters occurred after the divergence of bacteria and archaea. After performing over 60 replicate tree runs, we found that the best Firmicutes tree yielded robust UFBoot2 values, ranging between 91% and 100% for the ExtH node (Fig. 1d). When we repeated the analysis for the Proteobacteria phylum, the best tree had 5–87% UFBoot2 values for the equivalent nodes, reflecting nonreproducible trees. Thus, we used the Firmicutes tree for all subsequent analyses.
The run yielding the best tree, as determined by log likelihood, was inspected for long branches, defined as >0.7 substitutions per site. These were removed and, in the second round of tree-searching with the final dataset of 1,153 sequences, the analysis was repeated with 254 independent tree search runs. The final tree had the best-fit model of LG + F + R14, chosen according to the Bayesian information criterion. Bootstrap values were determined by UFBoot2 using 1,000 replicates45. Ancestral sequences were reconstructed from the best tree using the IQ-TREE implementation77 based on Yang’s marginal reconstruction method82,83,84. The ML sequences were generated using custom Python scripts, either written manually or generated using ChatGPT. The final ML amino acid sequence for each node was constructed using the amino acid with the highest PP at every site. All tree figures were generated using TreeViewer85. Visual representations of sequence alignments and PPs were made using WebLogos86.
Protein expression and purification
Because poorly aligned loops and tails are not reconstructed in our APR analysis, we had to graft them in for expression constructs. We took comparable loops and tails from an extant sequence close to AncInt (KJS87745.1); the 3L4 has two lysines that were substituted to histidines (K105H;K109H) to prevent potential proteolysis. All protein constructs were codon-optimized for E. coli, synthesized (GenScript) and cloned into a modified pBAD vector containing a C-terminal thrombin cleavage site followed by either an ALFA peptide tag87 or a C-terminal thermostable GFP specialized for membrane proteins49. An additional Twin-Strep tag and a 10×His tag were added to the C terminus of these constructs, all separated by glycine–serine (GSSS) linkers. The GltPh construct used was the ‘CAT7’ construct, with seven histidines previously introduced for stability13. Vectors were transformed into E. coli BL21(DE3) or DH10B cells (New England Biolabs, C2527; Invitrogen, 12331013), cells were grown in 2×YT medium and expression was induced by adding 0.1% arabinose at 0.8–1.0 OD600. Proteins were expressed for 2–3 h at 37 °C. Membranes were purified as previously described for GltPh, with all steps being performed at 4 °C (ref. 3). Briefly, cells were broken using a high-pressure cell disruptor (Avestin) for three passes with cooled water continuously applied. The resulting lysate was spun at 17,400g for 15 min and the supernatant was spun at 186,000g for 1 h. The resulting membrane pellet was homogenized in 20 mM HEPES–NaOH pH 7.4, 10 mM EDTA and 10% sucrose. Finally, the slow and fast spins were repeated and the resulting pellet was frozen at −80 °C.
All protein purification steps were performed at 4–8 °C. Crude membranes were homogenized in 20 mM HEPES–NaOH pH 7.4, 200 mM NaCl and 1 mM monopotassium l-aspartate; solubilization was initiated by the addition of 40 mM n-dodecyl-β-d-maltopyranoside (DDM; Anatrace) and the solubilizing membranes were gently rocked for 2 h. Over the course of our experiments, we noticed that tags were being nonspecifically proteolyzed upon solubilization; to reduce this, we added 1 mM PMSF and 1 mM EDTA before adding DDM. After solubilization, samples were clarified by high-speed ultracentrifugation at 186,000g for 1 h.
For ancestral constructs, the supernatant was applied to a prepacked 1-ml Streptactin XT 4Flow high-capacity column (IBA Biosciences) at a constant flow rate of 0.5–1 ml min−1. The resin was washed until the ultraviolet absorbance at 280 nm (UV280) was flat, corresponding to 14 column volumes of 20 mM HEPES–NaOH pH 7.4, 200 mM NaCl, 1 mM l-aspartate and 1 mM DDM. Subsequently, the resin was eluted with the same buffer containing 50 mM biotin (IBA Biosciences) and proteins were concentrated.
For GltPh, the supernatant was applied to a 1-ml HisTrap HP (Cytiva) at a constant flow rate of 1 ml min−1. The resin was washed until the UV280 was flat, corresponding to 30 column volumes of 20 mM HEPES–NaOH pH 7.4, 200 mM NaCl, 1 mM l-aspartate, 40 mM imidazole and 1 mM DDM. Subsequently, the resin was eluted with the same buffer containing increased imidazole (250 mM) and proteins were concentrated.
Reconstitution into proteoliposomes
Unilamellar liposomes were prepared essentially as previously described3. Briefly, a 3:1 (w/w) ratio of E. coli polar lipids and egg phosphatidylcholine (Avanti Polar Lipids) was dried, hydrated in 50 mM HEPES–Tris pH 7.4 and 200 mM KCl at a final concentration of 5 mg ml−1, flash-frozen and stored at −80 °C. On the day of the reconstitution, liposomes were thawed, extruded through a 400-nm filter 13 times and destabilized with Triton X-100 using a 1:2 (w/w) ratio of Triton X-100 to lipid. For the counterflow and Na+-coupled uptake experiments, GFP-free proteins were purified by size-exclusion chromatography (SEC) using a Superdex 200 Increase 10/300 column (Cytiva) in 20 mM HEPES–NaOH pH 7.4, 200 mM NaCl, 1 mM l-aspartate and 1 mM DDM; proteins were subsequently concentrated using a 100-kDa-cutoff concentrator. Proteins in all comparative experiments (Fig. 2d–g) were purified and reconstituted alongside one another. To form proteoliposomes, the purified protein was mixed with destabilized liposomes (1:100, w/w) for 30 min; then, detergent was removed with BioBeads SM-2 (Bio-Rad) at 80 mg ml−1, which were applied twice at 22 °C for 2 h, once overnight at 4 °C and once more at 22 °C for 2 h.
Proteoliposome uptake experiments
After detergent removal with BioBeads, proteoliposomes underwent the same general procedure for buffer exchange and concentration, regardless of experiment. Proteoliposomes were exchanged into the appropriate buffer using a PD-10 column (Cytiva), flash-frozen in liquid N2, thawed, concentrated by ultracentrifugation (100,000g for 1 h), resuspended to a final concentration of 50 mg ml−1, flash-frozen and stored at −80 °C. On the day of the experiment, the proteoliposomes were thawed and extruded through a 400-nm filter 23 times. To minimize sample dilution in the extruder, the proteoliposome volume was no less than 100 µl. All subsequent experiments were performed at 30 °C.
For uptake experiments, liposomes were prepared as described above to a final concentration of 50 mg ml−1 in 20 mM HEPES–Tris pH 7.4, 200 mM KCl and 100 mM choline chloride. To initiate Na+-driven uptake in the presence of a negative membrane potential (−102 mV), liposomes were diluted 1:100 in 20 mM HEPES–Tris pH 7.4, 2 mM KCl, 198 mM choline chloride, 100 mM NaCl, 0.9 µM valinomycin and 1 µM l-aspartate. The l-aspartate was a mixture of cold monopotassium l-aspartate (Research Products International) and [3H]l-aspartate (PerkinElmer or American Radiolabeled Chemicals). For negative controls without the Na+ gradient, NaCl was replaced with choline chloride.
To stop the reactions, 200 µl of reaction mixture (100 µg of liposomes) was diluted in 2 ml of ice-cold quenching buffer (20 mM HEPES–Tris pH 7.4 and 200 mM lithium chloride). The quenched reaction was immediately filtered through a 0.22-µm filter membrane (Millipore Sigma) and washed three times with 2 ml of quenching buffer. Washed membranes were inserted into scintillation vials and the membranes were soaked overnight in 5 ml of Econo-Safe counting cocktail. Radioactivity in liposomes was measured using an LS6500 scintillation counter (Beckman Coulter). Technical replicates from an individual reconstitution were taken in duplicate and averaged and at least two separate reconstitutions were taken per point. Data were analyzed using GraphPad Prism 10.
Proteoliposome counterflow experiments (sodium-dependent)
For counterflow experiments, the proteoliposomes after BioBead detergent removal were similarly exchanged in 20 mM HEPES–Tris pH 7.4, 200 mM KCl and 100 mM choline chloride using PD-10 columns. The exchanged liposomes were then separated into tubes for sodium conditions and choline conditions. For sodium conditions, an equivalent buffer containing 100 mM sodium chloride instead of choline chloride was added after exchange so that the final sodium concentration was 10 mM and the final choline concentration was 90 mM. Choline conditions were treated similarly, with the original choline buffer instead of the sodium buffer. A final concentration of 0.5 mM monopotassium l-aspartate was added to both conditions. Finally, the liposomes were flash-frozen and thawed twice, concentrated by ultracentrifugation (100,000g for 1 h), resuspended in the same internal buffer at 50 mg ml−1, flash-frozen and stored at −80 °C as described in the uptake experiments.
On the day of the experiment, proteoliposomes were thawed and freshly extruded, as described in the uptake experiments. Counterflow was initiated by diluting the liposomes 1:100 in an external buffer equivalent to the internal buffer, except that the cold 0.5 mM l-aspartate was replaced by 0.5 µM [3H]l-aspartate (American Radiolabeled Chemicals). Negative controls had an additional 0.5 mM cold l-aspartate in the external buffer. Reactions proceeded for the indicated time and were stopped and measured as described in the uptake experiments. Data were analyzed using GraphPad Prism 10.
Proteoliposome counterflow experiments (pH-dependent)
Proteoliposomes after BioBead detergent removal were exchanged in 50 mM potassium phosphate (KH2PO4–K2HPO4) buffer plus 0.5 mM l-aspartate (pH 6, 7 or 8) by three rounds of resuspension, two freeze–thaw cycles and ultracentrifugation (100,000g for 1 h). These were performed so that the total internal buffer exchange was greater than 99.5%. After the final round, proteoliposomes were resuspended to a final concentration of 25 mg ml−1, flash-frozen and stored at −80 °C. On the day of the experiment, proteoliposomes were thawed and freshly extruded as described in the uptake experiments. Counterflow was performed as in the sodium-dependent counterflow experiments, except the dilution factor was 1:50 and the quenching buffer was 50 mM MES–KOH buffer pH 5.5. Data were analyzed using GraphPad Prism 10.
nanoDSF
All steps were performed on the same day of the protein solubilization and purification because we observed variance in results depending on the freshness of the protein. We opted to use GFP-tagged ancestral proteins because the Tm of the fused GFP is greater than 90 °C (ref. 49), which does not interfere with the Tm values observed in our experiments. After affinity purification, the proteins with C-terminal GFP were concentrated using a 100-kDa-cutoff concentrator (MilliporeSigma). To remove ligands, the protein was dialyzed using a PD MiniTrap G-25 (Cytiva). For AncNa and AncInt P295273S;Q327305N, this buffer was 20 mM HEPES, 200 mM NMDG-Cl pH 7.4 and 1 mM DDM; for AncInt, this buffer was 50 mM KH2PO4–K2HPO4 pH 7 and 1 mM DDM. Different buffers were used for optimal first-derivative curves in ligand-free conditions and we observed identical Tm values in substrate-bound conditions regardless of dialysis buffer composition. The dialyzed, ligand-free protein was concentrated and diluted into the same respective buffers at a final concentration of ~1 mg ml−1; for AncInt, the buffer contained 10 mM decyl-maltoside (DM) instead of 1 mM DDM. This was necessary to observe melting of AncInt, presumably by the shorter chain detergent destabilizing the overall protein. Thermal stability in the presence and absence of various potential ligands was measured using a Tycho NT.6 instrument (Nanotemper) and individual Tm values were calculated by determining the appropriate local minimum in the first derivative of the 350-nm:330-nm ratio. Data were analyzed using GraphPad Prism 10.
MST
After affinity purification, the proteins with C-terminal GFP were concentrated using a 100-kDa-cutoff concentrator and then SEC-purified on a Superose 6 Increase 10/300 column (Cytiva) in 50 mM KH2PO4–K2HPO4 buffer pH 7 and 1 mM DDM. For the MST experiment, monopotassium l-aspartate was serially diluted twofold in water for a total of 16 concentrations. Each aspartate dilution was mixed with in a 1:1 (v/v) ratio of protein samples so that the final protein concentration was 100–300 nM, the appropriate salt was 100 mM, the DDM was 0.4 mM and the KH2PO4–K2HPO4 was 10–20 mM. Measurements were taken using Monolith NT.115 Blue/Red MST (Nanotemper), high MST power, a blue detector (for GFP) and premium capillaries (Nanotemper).
For data analysis, obvious outliers caused by aggregates (as determined by instrument software, MO.Control 2) were discarded. The ideal response time for each individual experiment was determined by instrument software and this was always between 1.5–5 s. Individual binding titrations were normalized to fraction bound by KD fits in MO.Affinity Analysis version 2.3. To obtain the final binding affinity value, the experiments were repeated three independent times, the data points were averaged and the KD was calculated from the average values. Data were analyzed using GraphPad Prism 10.
Cryo-EM sample preparation
Membrane scaffold protein (MSP1E3) was expressed and purified as previously described88 in the absence of Na+ or l-aspartate. A 3:1 (w/w) ratio of E. coli polar lipids and egg phosphatidylcholine (Avanti Polar Lipids) was dried, hydrated in 50 mM HEPES–Tris pH 7.4 and 200 mM KCl and solubilized in 72 mM DDM at final lipid concentration of 18 mM as previously described15,88.
For AncInt, GFP-free transporter constructs were affinity-purified, concentrated and mixed with MSP1E3 and solubilized lipids (both containing no Na+ or l-aspartate) at a molar ratio of 1.5:1:50, aiming for a final concentration of lipid greater than 5 mM. This mixture was incubated at 22 °C for 30 min; then, 350 mg of BioBeads SM-2 (Bio-Rad) per ml of mixture (w/v) was added for 2 h at 22 °C with gentle shaking and another round of BioBeads was applied overnight at 4 °C with gentle shaking. The resulting MSP1E3 and protein preparations were cleared by ultracentrifugation at 100,000g for 30 min. The supernatant was applied to a Superose 6 Increase 10/300 column in 20 mM HEPES pH 7.4, 100 mM NMDG-Cl and the peak fractions were concentrated to 7–9 mg ml−1, as measured by absorbance at 280 nm (A280). For the AncInt substrate-bound dataset, 1 mM l-aspartate was added to the sample just before freezing. Reconstitution efficiency was verified using SDS–PAGE.
GltPh was reconstituted into MSP1E3 as previously described15. Affinity-purified protein was further purified by SEC (Superdex 200 Increase, 10/300). Peak fractions were concentrated, mixed with MSP1E3 and solubilized lipids at a molar ratio of 0.75:1:50, aiming for a final concentration of lipid greater than 5 mM. This mixture was incubated and treated with BioBeads SM-2 in the exact same fashion as ancestral protein. The resulting MSP1E3 and protein preparations were exchanged in Na+-free buffer by three rounds of tenfold dilution and concentration and cleared by ultracentrifugation at 100,000g for 30 min. The supernatant was applied to a Superose 6 Increase 10/300 column in 20 mM HEPES–Tris pH 7.4 and 50 mM choline chloride, with the goal of mimicking previously published conditions. The peak fractions were concentrated to 8 mg ml−1, as measured by A280. Reconstitution efficiency was verified using SDS–PAGE.
To improve particle distribution, fluorinated Fos-Choline-8 (Anatrace) dissolved in water was added to the sample to a final concentration of 1.5 mM. UltrAuFoil R1.2/1.3 300-mesh gold grids (Quantifoil) were glow-discharged at 25 mA for 80 s. Then, 3 µl of protein was applied to each grid, incubated for 20 s under 100% humidity at 4 °C, blotted for 2.5 s under 0 blot force and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific).
Cryo-EM data acquisition and processing
Imaging was performed on a Titan Krios (FEI) equipped with a K3 camera (Gatan) using Leginon software. Specifics of each data collection and each individual map and model are provided in Table 1. Visual summaries of processing workflows and EM validation are provided in Extended Data Fig. 6 and Supplementary Figs. 4–6 and 8.
Initial processing pipeline for AncInt
For all datasets, raw videos were imported into cryoSPARC 4.0–4.4 and subjected to patch motion correction and patch contrast transfer function (CTF) estimation, where the videos were binned twice89. Low-quality videos were removed, as determined by CTF fits, ice thickness, intensity, etc. Particles were manually picked to train Topaz90, where, for each dataset, the optimal picking model and estimated number of particles per micrograph were empirically determined using the Topaz cross-validation tool in cryoSPARC. After picking and extraction with four binning rounds, an ab initio 3D model was constructed and high-quality particles were sorted using multiple heterogeneous refinement jobs, where ‘good’ particles sorted into the ab initio model class and ‘junk’ particles sorted into three pure noise ‘decoy’ volume classes (created by performing one iteration of ab initio). When greater than 95% of particles converged into the protein volume, the remaining particles were re-extracted to full box size and subjected to 3D classification without alignment and nonuniform (NU) refinement91. To maximize the number of particles obtained, the Topaz model was retrained on the clean particle stack after rounds of two-dimensional classification and heterogeneous refinement was repeated with the new particle stack and the high-resolution volume from NU refinement (replacing the ab initio volume). Remaining particles were re-extracted to full box size and subjected to NU refinement.
AncInt ligand-free dataset processing
Our ancestral protein datasets were highly heterogeneous and poorly symmetric; throughout the processing, we realized that they were reminiscent of other seemingly mobile membrane protein structures92,93. To overcome this, we attempted to enrich particles with symmetric features so that we could classify individual protomers. In the ligand-free dataset, we converted particle coordinates to RELION format using PyEM94. Particles were re-extracted in RELION using videos motion-corrected and binned twice using RELION’s implementation of MotionCor2 (ref. 95). These particles were reimported into cryoSPARC and refined using NU refinement (C1); the resulting particle images were converted to RELION format with PyEM. These particles were then subjected to two rounds of Bayesian polishing96, classification with heterogeneous refinement and classification with NU refinement (C1). To remove ‘disordered’ particles, we used 3D classification without alignment (C1) in cryoSPARC. We further optimized symmetric particles by performing one round of ab initio with C3 symmetry imposed. Using the best class, we achieved a resolution of 2.8 Å in C3, which allowed us to resolve individual protomers using symmetry expansion and focused classification using a mask on the transport domain. Each class underwent local refinement masked on the entire protomer (including transport and scaffold domains) using pose and shift Gaussian priors during alignment (standard deviations of 3° and 2 Å).
AncInt substrate-bound processing
As with the ligand-free dataset, the substrate-bound dataset was similarly heterogeneous and asymmetric. Using the previous approach, we encountered trouble imposing C3 symmetry, preventing focused classification of individual protomers. During our processing, we realized that we could more efficiently remove disordered particles using outputs from 3D variability (3DVA), where the first variability component reflects this ‘order–disorder’ transition between rigid, easily aligned particles and presumably mobile, difficult-to-align particles. We then took the first frame of the 3DVA video (disordered, bad volume), the last frame of the video (ordered, good volume) and two decoy noise volumes as inputs for heterogeneous refinement. Though a fundamentally similar approach to 3D classification used in the ligand-free dataset, the new approach allowed us to impose C3 symmetry with good resolutions (3.4 Å). We then symmetry-expanded the particles, performed 3D classification without alignment on the masked transport domain and locally refined the protomers using pose and shift Gaussian priors during alignment (standard deviations of 3° and 2 Å), as with the ligand-free dataset.
GltPh ligand-free processing
Raw videos were imported into cryoSPARC 4.4 and subjected to patch motion correction and patch CTF estimation, where the videos were binned twice89. Low-quality videos were removed, as determined by CTF fits, ice thickness, intensity, etc. Particles were manually picked to train Topaz90, where the optimal picking model and estimated number of particles per micrograph were empirically determined using the Topaz cross-validation tool in cryoSPARC. After picking and extraction, an ab initio 3D model was constructed. After one round of heterogeneous refinement with ab initio volumes, we performed NU refinement (C3) on the remaining particles, symmetry-expanded the particles, performed 3D classification without alignment on the masked transport domain and locally refined the protomers using pose and shift Gaussian priors during alignment (standard deviations of 3° and 2 Å).
AncInt P295273S;Q327305N ligand-free dataset processing
Because of the substantially lower resolution of the dataset, previously described methods to improve the resolution after decoy classification (3DVA and 3D classification without alignment) did not work. Therefore, we immediately performed 3D classification (C3) after decoy classification. We then performed symmetry expansion and 3D classification without alignment on the masked transport domain.
Model building and refinement
For model building, maps were sharpened using DeepEMhancer97. We generated a homology model of AncInt in SWISS-MODEL, using an outward-facing state of GltPh (PDB 2NWX) as a template13,98. The template for GltPh was PDB 6UWF (outward-facing) or PDB 6UWL (intermediate outward-facing). The model was adjusted into the density using ISOLDE99 and the final model was iteratively real-space refined and validated in PHENIX using the unsharpened density and default parameters100. All structural figures were made with ChimeraX101.
Gene neighborhood analysis
For each clade (ExtNa, ExtInt and ExtH), a sequence similarity network and corresponding genome neighborhood was created using the Enzyme Function Initiative webserver102. Transporters without information regarding protein-coding gene neighbors were excluded. A transporter was considered paired to an enzyme if they were within three neighboring genes away from the transporter. Genes were considered to have amino-acid-modifying or dicarboxylate-modifying activity according to Pfam annotation.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Atomic coordinates and EM densities were deposited to the PDB and EM Data Bank for the following structures: ligand-free AncInt low-affinity (PDB 9BGY, EMD-44526); ligand-free AncInt high-affinity (PDB 9BGZ, EMD-44527); l-aspartate-bound AncInt (PDB 9BH0, EMD-44528); ligand-free GltPh intermediate outward-facing (PDB 9BH1, EMD-44529); ligand-free GltPh outward-facing (PDB 9BH2, EMD-44530). Supplementary Data 2 includes all input and output data related to phylogenetic analyses, plasmid maps of all protein expression constructs used in this study and a ChimeraX session file containing comparison of AncInt models to AncInt P295273S;Q327305N maps (Extended Data Figure 9). Plasmid backbones designed in this study are available on Addgene (pBAD24-ALFA, 240248; pBAD24-TGP, 240249; pET15b-ALFA, 240252; pET15b-TGP, 240251). Previously published structural models are available from the PDB under accession codes 2NWX, 6UWF, 6X12, 6X15, 7AHK, 7RCP, 8CUI and 8CV2. All other plasmids and data are available upon request. Source data are provided with this paper.
References
Ryan, R. M., Compton, E. L. R. & Mindell, J. A. Functional characterization of a Na+-dependent aspartate transporter from Pyrococcus horikoshii. J. Biol. Chem. 284, 17540–17548 (2009).
Gaillard, I., Slotboom, D. J., Knol, J., Lolkema, J. S. & Konings, W. N. Purification and reconstitution of the glutamate carrier GltT of the thermophilic bacterium Bacillus stearothermophilus. Biochemistry 35, 6150–6156 (1996).
Reddy, K. D., Ciftci, D., Scopelliti, A. J. & Boudker, O. The archaeal glutamate transporter homologue GltPh shows heterogeneous substrate binding. J. Gen. Physiol. 154, e202213131 (2022).
Wheeler, L. C., Lim, S. A., Marqusee, S. & Harms, M. J. The thermostability and specificity of ancient proteins. Curr. Opin. Struct. Biol. 38, 37–43 (2016).
Trinco, G. et al. Kinetic mechanism of Na+-coupled aspartate transport catalyzed by GltTk. Commun. Biol. 4, 1–11 (2021).
Groeneveld, M. & Slotboom, D.-J. Na+:aspartate coupling stoichiometry in the glutamate transporter homologue GltPh. Biochemistry 49, 3511–3513 (2010).
Zerangue, N. & Kavanaugh, M. P. Flux coupling in a neuronal glutamate transporter. Nature 383, 634–637 (1996).
Guskov, A., Jensen, S., Faustino, I., Marrink, S. J. & Slotboom, D. J. Coupled binding mechanism of three sodium ions and aspartate in the glutamate transporter homologue GltTk. Nat. Commun. 7, 13420 (2016).
Verdon, G., Oh, S., Serio, R. N. & Boudker, O. Coupled ion binding and structural transitions along the transport cycle of glutamate transporters. eLife 3, e02283 (2014).
Boudker, O., Ryan, R. M., Yernool, D., Shimamoto, K. & Gouaux, E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature 445, 387–393 (2007).
Oh, S. & Boudker, O. Kinetic mechanism of coupled binding in sodium–aspartate symporter GltPh. eLife 7, e37291 (2018).
Reyes, N., Oh, S. & Boudker, O. Binding thermodynamics of a glutamate transporter homolog. Nat. Struct. Mol. Biol. 20, 634–640 (2013).
Yernool, D., Boudker, O., Jin, Y. & Gouaux, E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 431, 811–818 (2004).
Reyes, N., Ginter, C. & Boudker, O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature 462, 880–885 (2009).
Wang, X. & Boudker, O. Large domain movements through the lipid bilayer mediate substrate release and inhibition of glutamate transporters. eLife 9, e58417 (2020).
Arkhipova, V., Guskov, A. & Slotboom, D. J. Structural ensemble of a glutamate transporter homologue in lipid nanodisc environment. Nat. Commun. 11, 998 (2020).
Crisman, T. J., Qu, S., Kanner, B. I. & Forrest, L. R. Inward-facing conformation of glutamate transporters as revealed by their inverted-topology structural repeats. Proc. Natl Acad. Sci. USA 106, 20752–20757 (2009).
Ruan, Y. et al. Direct visualization of glutamate transporter elevator mechanism by high-speed AFM. Proc. Natl Acad. Sci. USA 114, 1584–1588 (2017).
Akyuz, N., Altman, R. B., Blanchard, S. C. & Boudker, O. Transport dynamics in a glutamate transporter homologue. Nature 502, 114–118 (2013).
Erkens, G. B., Hänelt, I., Goudsmits, J. M. H., Slotboom, D. J. & van Oijen, A. M. Unsynchronised subunit motion in single trimeric sodium-coupled aspartate transporters. Nature 502, 119–123 (2013).
Huang, Y. et al. Use of paramagnetic 19F NMR to monitor domain movement in a glutamate transporter homolog. Nat. Chem. Biol. 16, 1006–1012 (2020).
Grewer, C. et al. Individual subunits of the glutamate transporter EAAC1 homotrimer function independently of each other. Biochemistry 44, 11913–11923 (2005).
Groeneveld, M. & Slotboom, D.-J. Rigidity of the subunit interfaces of the trimeric glutamate transporter GltT during translocation. J. Mol. Biol. 372, 565–570 (2007).
Yernool, D., Boudker, O., Folta-Stogniew, E. & Gouaux, E. Trimeric subunit stoichiometry of the glutamate transporters from Bacillus caldotenax and Bacillus stearothermophilus. Biochemistry 42, 12981–12988 (2003).
Raunser, S. et al. Structure and function of prokaryotic glutamate transporters from Escherichia coli and Pyrococcus horikoshii. Biochemistry 45, 12796–12805 (2006).
Canul-Tec, J. C. et al. Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature 544, 446–451 (2017).
Gendreau, S. et al. A trimeric quaternary structure is conserved in bacterial and human glutamate transporters. J. Biol. Chem. 279, 39505–39512 (2004).
Alleva, C. et al. Na+-dependent gate dynamics and electrostatic attraction ensure substrate coupling in glutamate transporters. Sci. Adv. 6, eaba9854 (2020).
Bastug, T. et al. Position of the third Na+ site in the aspartate transporter GltPh and the human glutamate transporter, EAAT1. PLoS ONE 7, e33058 (2012).
Qiu, B. & Boudker, O. Symport and antiport mechanisms of human glutamate transporters. Nat. Commun. 14, 2579 (2023).
Qiu, B., Matthies, D., Fortea, E., Yu, Z. & Boudker, O. Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport. Sci. Adv. 7, eabf5814 (2021).
Garaeva, A. A., Guskov, A., Slotboom, D. J. & Paulino, C. A one-gate elevator mechanism for the human neutral amino acid transporter ASCT2. Nat. Commun. 10, 3427 (2019).
Tao, Z., Zhang, Z. & Grewer, C. Neutralization of the aspartic acid residue Asp-367, but not Asp-454, inhibits binding of Na+ to the glutamate-free form and cycling of the glutamate transporter EAAC1. J. Biol. Chem. 281, 10263–10272 (2006).
Riederer, E. A., Moënne-Loccoz, P. & Valiyaveetil, F. I. Distinct roles of the Na+ binding sites in the allosteric coupling mechanism of the glutamate transporter homolog, GltPh. Proc. Natl Acad. Sci. USA 119, e2121653119 (2022).
Riederer, E. A. & Valiyaveetil, F. I. Investigation of the allosteric coupling mechanism in a glutamate transporter homolog via unnatural amino acid mutagenesis. Proc. Natl Acad. Sci. USA 116, 15939–15946 (2019).
Zhou, W., Trinco, G., Slotboom, D. J., Forrest, L. R. & Faraldo-Gómez, J. D. On the role of a conserved methionine in the Na+-coupling mechanism of a neurotransmitter transporter homolog. Neurochem Res. 47, 163–175 (2022).
Starr, T. N. & Thornton, J. W. Epistasis in protein evolution. Protein Sci. 25, 1204–1218 (2016).
Wilson, C. et al.Using ancient protein kinases to unravel a modern cancer drug’s mechanism. Science 347, 882–886 (2015).
Ugalde, J. A., Chang, B. S. W. & Matz, M. V. Evolution of coral pigments recreated. Science 305, 1433–1433 (2004).
Harms, M. J. & Thornton, J. W. Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature 512, 203–207 (2014).
Gaucher, E. A., Govindarajan, S. & Ganesh, O. K. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 451, 704–707 (2008).
Pillai, A. S. et al. Origin of complexity in haemoglobin evolution. Nature 581, 480–485 (2020).
Lim, S. A., Hart, K. M., Harms, M. J. & Marqusee, S. Evolutionary trend toward kinetic stability in the folding trajectory of RNases H. Proc. Natl Acad. Sci. 113, 13045–13050 (2016).
Hochberg, G. K. A. & Thornton, J. W. Reconstructing ancient proteins to understand the causes of structure and function. Annu. Rev. Biophys. 46, 247–269 (2017).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Eick, G. N., Bridgham, J. T., Anderson, D. P., Harms, M. J. & Thornton, J. W. Robustness of reconstructed ancestral protein functions to statistical uncertainty. Mol. Biol. Evol. 34, 247–261 (2017).
Orlandi, K. N., Phillips, S. R., Sailer, Z. R., Harman, J. L. & Harms, M. J. Topiary: pruning the manual labor from ancestral sequence reconstruction. Protein Sci. 32, e4551 (2023).
Mulligan, C. & Mindell, J. A. Mechanism of transport modulation by an extracellular loop in an archaeal excitatory amino acid transporter (EAAT) homolog. J. Biol. Chem. 288, 35266–35276 (2013).
Cai, H. et al. An improved fluorescent tag and its nanobodies for membrane protein expression, stability assay, and purification. Commun. Biol. 3, 753 (2020).
Groeneveld, M., Weme, R. G. J. D. O., Duurkens, R. H. & Slotboom, D. J. Biochemical characterization of the C4-dicarboxylate transporter DctA from Bacillus subtilis. J. Bacteriol. 192, 2900–2907 (2010).
Majd, H. et al. Screening of candidate substrates and coupling ions of transporters by thermostability shift assays. eLife 7, e38821 (2018).
Nji, E., Chatzikyriakidou, Y., Landreh, M. & Drew, D. An engineered thermal-shift screen reveals specific lipid preferences of eukaryotic and prokaryotic membrane proteins. Nat. Commun. 9, 4253 (2018).
Seidel, S. A. I. et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–315 (2013).
Ewers, D., Becher, T., Machtens, J.-P., Weyand, I. & Fahlke, C. Induced fit substrate binding to an archeal glutamate transporter homologue. Proc. Natl Acad. Sci. USA 110, 12486–12491 (2013).
Hänelt, I., Jensen, S., Wunnicke, D. & Slotboom, D. J. Low affinity and slow Na+ binding precedes high affinity aspartate binding in the secondary-active transporter GltPh. J. Biol. Chem. 290, 15962–15972 (2015).
Jensen, S., Guskov, A., Rempel, S., Hänelt, I. & Slotboom, D. J. Crystal structure of a substrate-free aspartate transporter. Nat. Struct. Mol. Biol. 20, 1224–1226 (2013).
Verdon, G. & Boudker, O. Crystal structure of an asymmetric trimer of a bacterial glutamate transporter homolog. Nat. Struct. Mol. Biol. 19, 355–357 (2012).
Bendahan, A., Armon, A., Madani, N., Kavanaugh, M. P. & Kanner, B. I. Arginine 447 plays a pivotal role in substrate interactions in a neuronal glutamate transporter. J. Biol. Chem. 275, 37436–37442 (2000).
Scopelliti, A. J., Ryan, R. M. & Vandenberg, R. J. Molecular determinants for functional differences between alanine–serine–cysteine transporter 1 and other glutamate transporter family members. J. Biol. Chem. 288, 8250–8257 (2013).
Scopelliti, A. J., Font, J., Vandenberg, R. J., Boudker, O. & Ryan, R. M. Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5. Nat. Commun. 9, 38 (2018).
Ciftci, D. et al. Linking function to global and local dynamics in an elevator-type transporter. Proc. Natl Acad. Sci. USA 118, e2025520118 (2021).
Huysmans, G. H. M., Ciftci, D., Wang, X., Blanchard, S. C. & Boudker, O. The high-energy transition state of the glutamate transporter homologue GltPh. EMBO J. 40, e105415 (2021).
Seal, R. P. & Amara, S. G. A reentrant loop domain in the glutamate carrier EAAT1 participates in substrate binding and translocation. Neuron 21, 1487–1498 (1998).
Williams, P. D., Pollock, D. D., Blackburne, B. P. & Goldstein, R. A. Assessing the accuracy of ancestral protein reconstruction methods. PLOS Comput. Biol. 2, e69 (2006).
Motlagh, H. N., Wrabl, J. O., Li, J. & Hilser, V. J. The ensemble nature of allostery. Nature 508, 331–339 (2014).
Saavedra, H. G., Wrabl, J. O., Anderson, J. A., Li, J. & Hilser, V. J. Dynamic allostery can drive cold adaptation in enzymes. Nature 558, 324–328 (2018).
Sailer, Z. R. & Harms, M. J. Molecular ensembles make evolution unpredictable. Proc. Natl Acad. Sci. 114, 11938–11943 (2017).
Chisholm, L. O., Orlandi, K. N., Phillips, S. R., Shavlik, M. J. & Harms, M. J.Ancestral reconstruction and the evolution of protein energy landscapes. Annu. Rev. Biophys. 53, 127–146 (2024).
Kandasamy, P., Gyimesi, G., Kanai, Y. & Hediger, M. A. Amino acid transporters revisited: new views in health and disease. Trends Biochem. Sci. 43, 752–789 (2018).
Thorens, B. GLUT2, glucose sensing and glucose homeostasis. Diabetologia 58, 221–232 (2015).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Johnson, M. et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9 (2008).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Edgar, R. C. MUSCLE5: high-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 13, 6968 (2022).
Hoang, D. T. et al. MPBoot: fast phylogenetic maximum parsimony tree inference and bootstrap approximation. BMC Evolut. Biol. 18, 11 (2018).
Rozewicki, J., Li, S., Amada, K. M., Standley, D. M. & Katoh, K. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res 47, W5–W10 (2019).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Soubrier, J. et al. The influence of rate heterogeneity among sites on the time dependence of molecular rates. Mol. Biol. Evol. 29, 3345–3358 (2012).
Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 (2008).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018).
Yang, Z., Kumar, S. & Nei, M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141, 1641–1650 (1995).
Hanson-Smith, V., Kolaczkowski, B. & Thornton, J. W. Robustness of ancestral sequence reconstruction to phylogenetic uncertainty. Mol. Biol. Evol. 27, 1988–1999 (2010).
Koshi, J. M. & Goldstein, R. A. Probabilistic reconstruction of ancestral protein sequences. J. Mol. Evol. 42, 313–320 (1996).
Bianchini, G. & Sánchez-Baracaldo, P. TreeViewer: flexible, modular software to visualise and manipulate phylogenetic trees. Ecol. Evol. 14, e10873 (2024).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Götzke, H. et al. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 10, 4403 (2019).
Ritchie, T. K. et al. Chapter 11—reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Vallese, F. et al. Architecture of the human erythrocyte ankyrin-1 complex. Nat. Struct. Mol. Biol. 29, 706–718 (2022).
Paknejad, N., Sapuru, V. & Hite, R. K. Structural titration reveals Ca2+-dependent conformational landscape of the IP3 receptor. Nat. Commun. 14, 6897 (2023).
Asarnow, D., Palovcak, E. & Cheng, Y. asarnow/pyem: UCSF pyem v0.5. Zenodo https://doi.org/10.5281/zenodo.3576630 (2019).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 1–8 (2021).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 74, 519–530 (2018).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Oberg, N., Zallot, R. & Gerlt, J. A. EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme Function Initiative (EFI) web resource for genomic enzymology tools. J. Mol. Biol. 435, 168018 (2023).
Acknowledgements
We thank the following the people for their contributions: J. Thornton and members of the Thornton lab, M. Harms, L. Skrabanek and E. Fortea, for helpful discussions regarding bioinformatics and phylogenetics; members of the O.B. lab, especially A. Scopelliti, X. Wang, B. Qiu and Y. Lopez, for helpful discussions regarding biochemistry and cryo-EM; A. Accardi and C. Nimigean for critical reading of the manuscript; W. Eng for assistance with protein expression; GenScript for prompt and patient support with gene synthesis; V. Braband, B. Milorey and D. Leyva from NanoTemper for help with troubleshooting MST parameters; A. Frazier for input on the schematic figure. The work was supported by the National Institutes of Health (NIH) grants F32 NS102325 (to K.D.R.) and R37 NS134865 (to O.B.). O.B. is an investigator of the Howard Hughes Medical Institute. Phylogenetic analyses were performed on the Weill Cornell high-performance computing cluster maintained by the Scientific Computing Unit. ChatGPT was used to write some of the Python scripts involved in bioinformatic data analysis. All cryo-EM grids were initially screened at the Weill Cornell Cryo-EM Core Facility with help from C. Fluck. Ligand-free AncInt was collected at the New York University cryo-EM facility with help from B. Wang. Substrate-bound AncInt was collected at the Columbia University Medical Center Cryo-EM core with assistance from R. Grassucci. An earlier substrate-bound AncInt dataset (with assistance from K. Nehelu), the ligand-free GltPh dataset (with assistance from A. Owji and A. Ishemgulova) and the ligand-free AncInt P295273S;Q327305N dataset (with assistance from G. Angulli and C. Hernandez) were collected at the Simons EM Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (349247), NYSTAR and the NIH National Institute of General Medical Sciences (GM103310), with additional support from Agouron Institute (F00316) and NIH S10 OD019994-01.
Author information
Authors and Affiliations
Contributions
K.D.R. and O.B. conceptualized the project and wrote the manuscript. K.D.R. performed all evolutionary analyses. K.D.R., B.R. and F.B.A. performed the functional experiments, prepared the grids and processed the cryo-EM data. K.D.R. and B.R. prepared the figures. K.D.R., B.R., F.B.A. and O.B. built and refined the molecular models. N.K. and S.P., under the supervision of K.D.R., performed the foundational pilot experiments, including expression and purification optimization, and preliminary cryo-EM data collection and processing. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Rachelle Gaudet and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Katarzyna Ciazynska, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Elevator mechanism of SLC1-type transporters.
GltPh structures in the substrate-bound outward-facing state (OFS-Asp, PDB 6UWF) and substrate-free inward-facing open state (IFS ligand-free, 6X12). The rigid scaffold domain is in transparent wheat; the mobile transport domain is in blue; HP1 is in yellow; HP2 is in red; purple spheres are Na+; green molecule is L-aspartate. In a typical transport cycle, three Na+ ions and L-aspartate bind in the outward-facing state, resulting in the closure of the HP2 gate and translocation of the transport domain to the inward-facing state. The HP2 gate then opens, releasing solutes to the intracellular side.
Extended Data Fig. 2 APR workflow to capture ExtNa to ExtH evolutionary transition.
The diagram is based on Orlandi et al. (1) Based on previous biochemical and structural characterization of prokaryotic H+-coupled (orange; GltPEc, GltTBc, DctABs) and Na+-coupled (purple; GltPh, GltTk) transporters, we identified signature residues that helped us sort putative sequences as either ExtH or ExtNa. (2) We retrieved homologs of transporters using PSI-BLAST. After clustering and quick sequence alignment, we removed sequences with large gaps or insertions. Finally, we used parsimony tree generation to keep sequences that encompass ExtH, ExtNa, and intermediate (green) transporters. (3) We carefully aligned the sequences using MAFFT-DASH, which incorporates structural information. Sequences with large gaps or insertions were removed. The remaining sequences were re-aligned, and poorly aligned loops and tails were stripped from the alignment. (4) The final sequence alignment was used as input for ML tree generation, using archaeal GltPh as the root. The optimal model of protein evolution was selected using ModelFinder. Sequences that caused long branches were removed, and 254 parallel tree searches were performed to find the optimal branch lengths and topology, resulting in the ML tree with the highest possible likelihood. (5) To reconstruct ancestral sequences, every node was reconstructed using optimized ML parameters and protein evolution model; each site in each node is assigned probabilities for all 20 amino acids, and the highest-probability amino acid at a site is selected for the ML sequence. (6) To determine if a given node is creating a robustly reconstructed ancestor, bootstrap is performed (UFBoot2), which calculates branch supports by resampling the MSA 1000 times and determining the frequency of the branch re-occurring in the trees. Sequence sampling is critical in this step; when we retrieved sequences from Firmicutes/Bacillota, we obtained robust UFBoot2 values for AncInt and descendants, but not when retrieving sequences from Proteobacteria. (7) Ancestral sequences are engineered for expression and purification by inserting loops and tails from extant sequences (green), which were not considered during APR due to poor alignment. All relevant references here and elsewhere in the legends are in the Main Text or Methods. Created in BioRender. Boudker, O. (2025) https://BioRender.com/sl3la84.
Extended Data Fig. 3 Phylogeny, rooting, and conservation of prokaryotic glutamate transporter homologs.
(a) Maximum parsimony phylogeny of bacterial glutamate transporter homologs. The clade containing conserved signature residues in H+-coupled transporters is colored orange. (b) Selection of initial sequences for APR analysis from Firmicutes/Bacillota (99% clustered). Purple, green, and orange clades contain representative ExtNa, ExtInt, and ExtH sequences, respectively. Gray sequences were unassigned in our analysis, and were eventually removed due to potential long-branch attraction artifacts. (c) Conservation of Na+- and H+-coupling signature residues in non-redundant archaeal glutamate transporters. (d) Conservation of the Na+-binding sites throughout the ML phylogeny. Top: sequences of Na+-coupled GltPh and H+-coupled GltTBc encompassing Na+-binding sites of ExtNa. The numbers above the sequences mark side chains in GltPh that coordinate Na+ sites 1–3; A indicates side chains coordinating L-aspartate. Bottom: WebLogos of the relative amino acid frequencies in ExtNa, ExtInt, and ExtH clades. The number of sequences per clade is 186, 161, and 786, respectively.
Extended Data Fig. 4 Robustness of the ancestral protein reconstruction.
(a) Reconstructed ancestral sequences in Na+-binding sites of AncNa, AncInt, and AncH. WebLogos visually represent the posterior probability per site in each ancestral sequence. The topmost residue at each site is the ML residue. GltPh and GltTBc sequence alignments are shown above the WebLogos for reference, along with annotations for L-aspartate binding residues (A) or Na+-binding residues1,2,3. (b) Average posterior probability (PP) per site, the natural logarithm of the total posterior probability of the entire ancestral sequence (Ln PP), and the fraction of the ambiguous sites (defined as two or more amino acids per site with a probability greater than 0.2) in each ancestor. Bold, larger values are for the entire sequence; values in parentheses are for the transport domain alone. (c) Pairwise identity (in %) matrix of ancestral and extant transporters. The alignment included only transport domains, defined as GltPh residues 78–110, 228–416. (d) Visual representation of the changes between two ML ancestral sequences. In each bar, the first segment is the number of changes where the posterior probability is greater than 0.9 in only the first ancestor, that is, a robustly reconstructed site is ambiguously reconstructed in the second ancestor; the second segment is the number of changes where the posterior probability is greater than 0.9 in only the second ancestor, while the site is ambiguous in the first ancestor; the third segment is the number of changes where the posterior probability is greater than 0.9 in both ancestors with both sites confidently reconstructed. (e, f) Probable sequence changes between AncNa and AncInt (e) and AncInt and AncH (f), mapped onto the transport domain of GltPh. Bottom: Labeling scheme for residues. Larger spheres represent sequence transitions; smaller spheres represent Na+ sites and structured waters observed in GltPh (PDB 7RCP). ‘Unknown’ indicates when there is no clear physicochemical difference between the two residues. ExtH signature residues are [T/S]92[A/L/I/V/M], M311L, and D405N; putative protonation site is S279E.
Extended Data Fig. 5 Sequence alignment of extant and ancestral glutamate transporter homologs.
(a) Alignment sequence numbering corresponds to GltPh numbering. Extant sequences are GltPh (UniProt O59010), GltTBc (P24944), and human EAAT3 (P43005). TMs, shown above the alignment, are colored wheat for the scaffold/trimerization domain, blue for the transport domain, yellow for re-entrant helical hairpin HP1, and red for re-entrant helical hairpin HP2. Dashed green lines represent residues grafted from an extant sequence into ancestral protein constructs in place of poorly aligned regions (see Methods). Filled purple circles mark residues in GltPh that coordinate Na+ ions with their side chains, filled green squares mark residues in GltPh that coordinate L-aspartate with their side chains, and empty circles represent allosteric mutations that can turn Na+-coupling on and off. Notable sequence changes in H+-coupled transporters compared to Na+-coupled transporters are shown in bold, and red residues are changes found at Na+-binding sites. (b) Structure of a GltPh protomer in the outward-facing state (PDB 6UWF), colored as in (a). Purple spheres are Na+ ions (placed based on PDB 6×15), and the substrate is green.
Extended Data Fig. 6 Validation statistics for all structures.
Map validations of (a) ligand-free AncInt, low-affinity; (b) ligand-free AncInt, high-affinity; (c) L-aspartate-bound AncInt; (d) ligand-free GltPh, low-affinity, iOFS; (e) ligand-free GltPh, low-affinity, OFS. From left to right, map FSC from local refinement in cryoSPARC and local resolution estimation of the unsharpened map. All maps are contoured at 8 σ. The adjacent protomers (unused in refinement) are removed for clarity.
Extended Data Fig. 7 Global transport domain conformations of structures obtained in this study.
AncInt (green) or GltPh (purple) structures were aligned on the scaffold domain (residues 1–75, 140–225) of the closest GltPh structure in the PDB (translucent white). These GltPh structures were either OFS (2NWX) or iOFS (6UWL). IFS states in AncInt were incompletely resolved, and models could not be built; therefore, the maps from 3D classification were contoured to a σ of 6 and aligned to the closest GltPh structure, IFS-open (white, 6 × 12).
Extended Data Fig. 8 Alternate substrate binding configurations and AncInt HP2 densities.
(a) Substrate-binding sites of ligand-free iOFS GltPh in the low-affinity configuration. (b) Substrate-binding site of ligand-free AncInt in the high-affinity configuration #2. The green density overlaps with the L-aspartate binding site but is too large and differently shaped to correspond to L-aspartate. (c–e) HP2 densities of ligand-free AncInt, low-affinity (c), ligand-free AncInt, high-affinity (d), and L-aspartate AncInt, high-affinity (e). Densities are unsharpened and contoured 10 σ. HP1 is in yellow, and HP2 is in red.
Extended Data Fig. 9 Functional characterization of mutant ancestral proteins.
(a) nanoDSF assays of Anc*, AncNa S295273P/N327305Q, and single AncInt mutations. Melting temperatures (Tm) in buffer alone (ligand-free), Na+, L-aspartate, and Na+/L-aspartate. Replicates are from at least two independent experiments, each with several technical replicates. All ligand concentrations are 10 mM Na+ and 1 mM L-aspartate, unless otherwise indicated. (b) Alignment of all 20 classes of AncInt P295273S/Q327305N protomers (obtained by 3D classification without alignment, additional detail in Methods) to the scaffold domains of either AncInt low-affinity or high-affinity states.
Extended Data Fig. 10 Genomic pairing of uncoupled transporters and enzymes.
Transporters were considered to have a paired enzyme if the enzyme was within 3 protein-coding genes away from the transporter and if the enzyme had putative free amino acid/dicarboxylate modifying function according to Pfam annotation. The fraction of sequences with genomic transporter/enzyme pairs is on the top of each bar. Only transporters with available genomic neighborhood information were considered; for the ExtNa, ExtInt, and ExtH clades, this corresponds to 29/186, 38/161, and 170/786, respectively.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Table 1 and Source data for Supplementary Figs. 1 and 3.
Supplementary Data 1
Uncropped Coomassie gels and Supplementary Figs. 1 and 3.
Supplementary Data 2
All input and output data related to phylogenetic analyses, plasmid maps of all protein expression constructs used in this study, a ChimeraX session file containing comparison of AncInt models to AncInt P295273S;Q327305N maps (Extended Data Figure 9), a summary table of all structural maps and models and PDB validation reports.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed gels.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Reddy, K.D., Rasool, B., Akher, F.B. et al. Evolutionary analysis reveals the origin of sodium coupling in glutamate transporters. Nat Struct Mol Biol 32, 2564–2574 (2025). https://doi.org/10.1038/s41594-025-01652-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01652-z
This article is cited by
-
A structural window into the evolution of secondary transport mechanisms
Nature Structural & Molecular Biology (2025)