Abstract
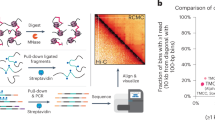
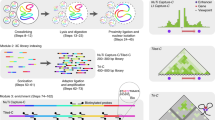
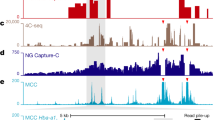
Micro Capture-C (MCC) is a chromatin conformation capture (3C) method for visualizing reproducible three-dimensional contacts of specified regions of the genome at base pair resolution. These methods are an established family of techniques that use proximity ligation to assay the topology of chromatin. MCC can generate data at substantially higher resolution than previous techniques through multiple refinements of the 3C method. Using a sequence agnostic nuclease, the maintenance of cellular integrity and full sequencing of the ligation junctions, MCC achieves subnucleosomal levels of resolution, which can be used to reveal transcription factor binding sites analogous to DNAse I footprinting. Gene dense regions, close-range enhancer–promoter contacts, individual enhancers within super-enhancers and multiple other types of loci or regulatory regions that were previously challenging to assay with conventional 3C techniques, are readily observed using MCC. MCC requires training in common molecular biology techniques and bioinformatics to perform the experiment and analyze the data. The protocol can be expected to be completed in a 3 week timeframe for experienced molecular biologists.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (GSE144336).
Code availability
The code required for analysis of MCC data are available for academic use through the Oxford University Innovation software store (https://process.innovation.ox.ac.uk/software/p/16529a/micro-capture-c-academic/1). A pipeline to run this code is available on GitHub (https://github.com/jojdavies/Micro-Capture-C). Instructions for setting up and running the pipeline are available on github.
References
Cavalli, G. & Misteli, T. Functional implications of genome topology. Nat. Struct. Mol. Biol. 20, 290–299 (2013).
Lam, M. T. Y., Li, W., Rosenfeld, M. G. & Glass, C. K. Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. https://doi.org/10.1016/j.tibs.2014.02.007 (2014).
Arnould, C. et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature 590, 660–665 (2021).
Marchal, C., Sima, J. & Gilbert, D. M. Control of DNA replication timing in the 3D genome. Nat. Rev. Mol. Cell Biol. 20, 721–737 (2019).
Jain, S., Ba, Z., Zhang, Y., Dai, H. Q. & Alt, F. W. CTCF-binding elements mediate accessibility of RAG substrates during chromatin scanning. Cell 174, 102–116.e14 (2018).
Hirota, T., Gerlich, D., Koch, B., Ellenberg, J. & Peters, J. M. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117, 6435–6445 (2004).
Lee, T. I. & Young, R. A. Transcriptional regulation and its misregulation in disease. Cell 152, 1237–1251 (2013).
Krumm, A. & Duan, Z. Understanding the 3D genome: emerging impacts on human disease. Semin. Cell Dev. Biol. 90, 62–77 (2019).
Cremer, T. & Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2, a003889 (2010).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Fortin, J. P. & Hansen, K. D. Reconstructing A/B compartments as revealed by Hi-C using long-range correlations in epigenetic data. Genome Biol. 16, 1–23 (2015).
Falk, M. et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570, 395–399 (2019).
S, S. & P, F. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019).
Hua, P. et al. Defining genome architecture at base-pair resolution. Nature 595, 125–129 (2021).
Aljahani, A. et al. Analysis of sub-kilobase chromatin topology reveals nano-scale regulatory interactions with variable dependence on cohesin and CTCF. Nat. Commun. 13, 2139 (2022).
Lakadamyali, M. & Cosma, M. P. Visualizing the genome in high resolution challenges our textbook understanding. Nat. Methods 17, 371–379 (2020).
Beagrie, R. A. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 (2017).
Davies, J. O. J., Oudelaar, A. M., Higgs, D. R. & Hughes, J. R. How best to identify chromosomal interactions: a comparison of approaches. Nat. Methods 14, 125–134 (2017).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Cairns, J. et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 17, 127 (2016).
Cullen, K. E., Kladde, M. P. & Seyfred, M. A. Interaction between transcription regulatory regions of prolactin chromatin. Science 261, 203–206 (1993).
Hagege, H. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 (2007).
Marbouty, M. et al. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell 59, 588–602 (2015).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Ma, W. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat. Methods 12, 71–78 (2014).
Ramani, V. et al. Mapping 3D genome architecture through in situ DNase Hi-C. Nat. Protoc. 11, 2104–2121 (2016).
Hsieh, T. H. S. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell 162, 108–119 (2015).
Hughes, J. R. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 46, 205–212 (2014).
Zhao, Z. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347 (2006).
Davies, J. O. J. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods 13, 74–80 (2016).
Downes, D. J. High-resolution targeted 3C interrogation of cis-regulatory element organization at genome-wide scale. Nat. Commun. 12, 531 (2021).
Downes, D. J. et al. Capture-C: a modular and flexible approach for high-resolution chromosome conformation capture. Nat. Protoc. 17, 445–475 (2022).
Akgol Oksuz, B. et al. Systematic evaluation of chromosome conformation capture assays. Nat. Methods 18, 1046–1055 (2021).
Mieczkowski, J. et al. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 7, 11485 (2016).
Krietenstein, N. et al. Ultrastructural details of mammalian chromosome architecture. Mol. Cell 78, 554–565.e7 (2020).
Galas, D. J. & Schmitz, A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5, 3157 (1978).
Aljahani, A. et al. Analysis of sub-kilobase chromatin topology reveals nano-scale regulatory interactions with variable dependence on cohesin and CTCF. Nat. Commun. 13, 2139 (2022).
Xie, T. et al. Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse. Genome Res. 13, 2621 (2003).
Nasser, J. et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021).
King, A. J. et al. Reactivation of a developmentally silenced embryonic globin gene. Nat. Commun. 12, 4439 (2021).
The Severe Covid-19 GWAS Group. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 383, 1522–1534 (2020).
Downes, D. J. et al. Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus. Nat. Genet. 53, 1606–1615 (2021).
Schmitt, A. D. et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 17, 2042–2059 (2016).
Giorgetti, L. et al. Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579 (2016).
van Bemmel, J. G. et al. The bipartite TAD organization of the X-inactivation center ensures opposing developmental regulation of Tsix and Xist. Nat. Genet. 51, 1024–1034 (2019).
Monahan, K., Horta, A. & Lomvardas, S. Lhx2/Ldb1-mediated trans interactions regulate olfactory receptor choice. Nature 565, 448 (2019).
Rao, S. S. P. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Li, G. Chromatin interaction analysis with paired-end tag (ChIA-PET) sequencing technology and application. BMC Genomics 15, S11 (2014).
Fang, R. et al. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 26, 1345–1348 (2016).
Mumbach, M. R. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016).
Schwartzman, O. UMI-4C for quantitative and targeted chromosomal contact profiling. Nat. Methods 13, 685–691 (2016).
Andrey, G. et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340, 1234167 (2013).
Smemo, S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371 (2014).
Ghavi-Helm, Y. et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512, 96–100 (2014).
Oudelaar, A. M. et al. Dynamics of the 4D genome during in vivo lineage specification and differentiation. Nat. Commun. 11, 2722 (2020).
Mifsud, B. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015).
Oudelaar, A. M., Downes, D., Davies, J. & Hughes, J. Low-input capture-C: a chromosome conformation capture assay to analyze chromatin architecture in small numbers of cells. Bio. Protoc. 7, e2645 (2017).
Beagrie, R. A. et al. Multiplex-GAM: genome-wide identification of chromatin contacts yields insights not captured by Hi-C. Preprint at bioRxiv https://doi.org/10.1101/2020.07.31.230284 (2020).
Winick-Ng, W. et al. Cell-type specialization is encoded by specific chromatin topologies. Nature 599, 684–691 (2021).
Quinodoz, S. A. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757.e24 (2018).
Takei, Y. et al. Integrated spatial genomics reveals global architecture of single nuclei. Nature 590, 344–350 (2021).
Tan, L., Xing, D., Chang, C. H., Li, H. & Xie, X. S. Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018).
Owens, D. D. G. et al. Dynamic Runx1 chromatin boundaries affect gene expression in hematopoietic development. Nat. Commun. 13, 773 (2022).
Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 9, 999–1003 (2012).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome https://doi.org/10.1186/s13059-015-0831-x (2015).
Buckle, A., Gilbert, N., Marenduzzo, D. & Brackley, C. A. capC-MAP: software for analysis of Capture-C data. Bioinformatics https://doi.org/10.1093/bioinformatics/btz480 (2019).
Thongjuea, S., Stadhouders, R., Grosveld, F. G., Soler, E. & Lenhard, B. R3Cseq: an R/Bioconductor package for the discovery of long-range genomic interactions from chromosome conformation capture and next-generation sequencing data. Nucleic Acids Res. 41, e132 (2013).
Smith, A. & Rue-Albrecht, K. sims-lab/CapCruncher. Zenodo https://doi.org/10.5281/zenodo.5113088 (2021).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Skene, P. J., Henikoff, J. G. & Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018).
Hentges, L. D., Sergeant, M. J., Downes, D. J., Hughes, J. R. & Taylor, S. LanceOtron: a deep learning peak caller for genome sequencing experiments. Bioinformatics 38, 4255–4263 (2022).
Oudelaar, A. M. et al. Single-allele chromatin interactions identify regulatory hubs in dynamic compartmentalized domains. Nat. Genet. 50, 1744–1751 (2018).
Schoenfelder, S. et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 25, 582–597 (2015).
Cheng, L. et al. Single-nucleotide-level mapping of DNA regulatory elements that control fetal hemoglobin expression. Nat. Genet. 53, 869–880 (2021).
Acknowledgements
J.O.J.D. was supported by grants from Wellcome (098931/Z/12/Z and 225220/Z/22/Z); the MRC (MR/R008108/1 and MC_UU_00029/04); the NIHR Blood and Transplant Research Unit (NIHR203339) and the Oxford Biomedical Research Centre Genomic Medicine and Cell and Gene Therapy Themes (NIHR203311). J.C.H. is supported by an MRC studentship and N.D. was supported by an NIHR Academic Clinical Fellowship (ACF-2019-13-013).
Author information
Authors and Affiliations
Contributions
J.O.J.D. developed the method and wrote the first draft of the manuscript. J.C.H., H.L. and N.D. optimized the method and wrote the manuscript. D.D. optimized the capture methodology and contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.O.J.D. is a co-founder of Nucleome Therapeutics and he provides consultancy to the company. He also holds a patent for the MCC method, which is licensed to the company.
Peer review
Peer review information
Nature Protocols thanks Argyris Papantonis and Vijay Ramani for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key papers using this protocol
Hua, P. et al. Nature 595, 125–129 (2021): https://doi.org/10.1038/s41586-021-03639-4
Aljahani, A. et al. Nat. Commun. 13, 2139 (2022): https://doi.org/10.1038/s41467-022-29696-5
Downes, D. J. et al. Nat. Genet. 53, 1606–1615 (2021): https://doi.org/10.1038/s41588-021-00955-3
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hamley, J.C., Li, H., Denny, N. et al. Determining chromatin architecture with Micro Capture-C. Nat Protoc 18, 1687–1711 (2023). https://doi.org/10.1038/s41596-023-00817-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-023-00817-8
This article is cited by
-
CiFi: accurate long-read chromosome conformation capture with low-input requirements
Nature Communications (2025)
-
CTCF depletion decouples enhancer-mediated gene activation from chromatin hub formation
Nature Structural & Molecular Biology (2025)
-
Chromatin interaction maps of human arterioles reveal mechanisms for the genetic regulation of blood pressure
Nature Communications (2025)
-
Enhancer Enh483 regulates myoblast proliferation and differentiation of buffalo myoblasts by targeting FAXC
Cell and Tissue Research (2025)
-
Predicting gene expression state and prioritizing putative enhancers using 5hmC signal
Genome Biology (2024)