Abstract
The Traditional Formula (TF), a combination of herbs prepared in accordance with traditional medicine principles, is increasingly garnering global attention as an alternative to modern medicine. Specifically, there is growing interest in exploring TF’s therapeutic effects across various diseases. A significant portion of the state-of-the-art knowledge regarding the relationship between TF and disease is found in scientific publications, where manual knowledge extraction is impractical. Thus, Natural Language Processing (NLP) is being employed to efficiently and accurately search and extract crucial knowledge from unstructured literatures. However, the absence of a high-quality manually annotated corpus focusing on TF-disease relationships hampers the use of NLP in the fields of traditional medicine and modern biomedical science. This article introduces the Traditional Formula-Disease Relationship (TFDR) corpus, a manually annotated corpus designed to facilitate the automatic extraction of TF-disease relationships from biomedical literatures. The TFDR corpus includes information gleaned from 740 PubMed abstracts, encompassing a total of 6,211 TF mentions, 7,166 disease mentions, and 1,109 relationships between them encapsulated within 744 key-sentences.
Similar content being viewed by others
Background & Summary
Advancements in science and technology have led to the accumulation of vast amounts of data in all fields. Much of this data exists in unstructured text format, which may contain innovative information that has not been previously revealed1. Unstructured text in the biomedical domain, in particular, is known to contain useful information related to drug development, drug repurposing, clinical application and more2. However, these diverse and massive text is increasing rapidly; thus, manually extracting and analyzing useful information from them is impossible and most significant information remains unstructured within the vast texts. As a result, natural language processing (NLP) is being utilized to efficiently and accurately search and extract crucial information from unstructured literatures. Recently, remarkable improvements have been achieved in the field of NLP, such as named entity recognition, relation extraction, and knowledge graph creation, by integrating deep learning-based technology3. The success of recent NLP research in the biomedical field can be attributed to the prior development of large, high-quality corpora, as well as advancements in computing power and deep learning techniques4.
Traditional medicine, with its rich clinical experience spanning thousands of years, is rooted in a holistic approach that seeks to restore balance within the body through its diagnostic and therapeutic practices5. A key element of traditional medicine systems is the Traditional Formula (TF), which is composed of a carefully crafted combination of herbs—a term that includes not only medicinal plants but also animal products and minerals. These formulas are designed according to traditional medical principles and holistic philosophies specific to systems like Traditional Chinese Medicine (TCM), Ayurveda, Unani, and others6. TFs are more than just simple mixtures; they are synergistic combinations meant to address both the underlying causes of illness and its symptoms. In many countries across Asia, Africa, and Latin America, TFs are widely accepted and used as a primary form of healthcare. In contrast, in countries like those in Europe, the United States, and Australia, TFs are often viewed as part of complementary and alternative medicine (CAM)7.
TF is increasingly attracting global attention as an alternative to modern medicine, and numerous researches have been published that clarifies the important mechanisms of action and therapeutic effects of TF in various diseases such as cancer, cardiovascular disease, and infections, using modern scientific methods8,9,10. The large-scale analysis of traditional medical literatures has become an interesting research field in recent years, providing the potential to generate new knowledge for activating clinical treatments, multi-drug pharmacology researches, and enhancing understanding of traditional medicine11. The application of NLP techniques holds great potential in extracting novel insights regarding the relationship between TF and diseases through extensive analysis of scientific literatures. However, the effectiveness of NLP tasks heavily relies on the availability of a comprehensive and well-structured corpus that encompasses the TF-disease relationship.
To facilitate the application of deep learning-based NLP tasks, ongoing endeavors within the biomedical and traditional medicine domains are focused on generating expansive and thoroughly curated corpora from biomedical literatures. These efforts aim to produce corpora of substantial size and exceptional quality, enabling enhanced research and analysis in the field of NLP. In the field of biomedicine, there exist corpora such as AIMed12, BioInfer13, POMELO14, Plant-Disease15, and PPR4, which have been created to elucidate diverse relationships among biomedical entities. While most of these publicly available datasets primarily focus on modern medicine, the availability of datasets for traditional medicine is relatively limited. The limited availability of corpora for traditional medicine is due to the complexity and variability within these systems, which often lack standardized terminology, making it more challenging to develop large-scale corpora like those in modern biomedical research.
Nevertheless, there are several manually curated corpora that describe diverse relationships among entities in traditional medicine. Examples includes DFI-DB16, FG-ERC17, MSM18, and StrokeKG19. DFI-DB, a comprehensive resource for drug-food interactions, was developed by annotating abstracts from PubMed, covering not only drug-food but also drug-herb interactions. FG-ERC, on the other hand, was created by annotating 13 types of TCM entities, such as acupuncture points, diagnostic methods, and TF, extracted from Chinese clinical records. MSM focuses on mixture symptom entities and their relationships, using annotations from clinical records written in Chinese. StrokeKG, automatically constructed from TCM-related abstracts in PubMed, captures nine entity types, including herbs and Chinese patent medicines (CPM), with relationships like herb-disease and CPM-disease specifically related to stroke. Despite the diversity of these corpora, none of them focus specifically on the systematic relationships between TF and diseases in biomedical publications written in English. For example, although FG-ERC includes TF as one of its entity types from Chinese clinical records, it does not emphasize TF and disease relationships. Similarly, while StrokeKG captures herb-disease relationships, its focus on stroke-related content limits its coverage of the broader scope of TF-disease connections.
In this study, we conducted manual annotation of 740 abstracts sourced from PubMed in order to develop the Traditional Formula-Disease Relationship (TFDR) corpus. The TFDR corpus comprises a total of 6,211 TF mentions and 7,166 disease mentions, along with 1,109 relationships between them, all of which are contained within 744 key-sentences, which are condensed representations of the result or conclusion of an article, containing both TF and disease mentions. To the best of our knowledge, the TFDR corpus represents the first of its kind. Furthermore, we evaluated the baseline performance of TFDR using various Transformer20 encoder models, which are well-known for their effectiveness in a wide range of NLP tasks.
Methods
Lexical resources
In order to construct a corpus that captures the relationship between TF and diseases described in biomedical publications, it is crucial to have access to comprehensive vocabularies for both TF and diseases. However, the descriptions of TF in scientific literature exhibit substantial variation across different authors, and there is currently a lack of a unified TF vocabulary that can accommodate such diversity. To address this challenge, we utilized a Traditional Korean Medicine (TKM) ontology21 that extracted a wide range of TKM terms, such as medicinal herbs, formulas, symptoms, and meridians. This ontology also established the interrelationships among these terms by drawing on information from authoritative TKM textbooks and classical medical texts. The TF vocabulary comprises 446 representative names and 922 synonyms. In order to gather a wide range of English transliterations for the TF, several databases including OASIS (https://oasis.kiom.re.kr/index.jsp), CNKI (https://oversea.cnki.net/index/), Kampo DB (https://wakanmoview.inm.u-toyama.ac.jp/kampo/), Chinese Medicine Formulae Image Database (https://sys02.lib.hkbu.edu.hk/cmfid/index.asp), and TCM Wiki (https://tcmwiki.com/) were utilized. These databases served as valuable resources for collecting diverse English expressions associated with the TF. Three TCM doctors conducted independent searches within databases and cross-verified the results during weekly meetings. If they were unable to reach a consensus on the outcomes during these meetings, a TKM doctor made the final decision to create the TF vocabulary. By adopting this approach, we successfully constructed a comprehensive TF vocabulary that encompasses the heterogeneous English expressions of TF encountered in scholarly literature. As an example, the traditional formula 五積散 has been assigned different English transliterations, including ojeok-san, wuji-san, goseki-san, and goshaku-san. By consolidating these transliterations, we performed a comprehensive search on PubMed to identify relevant articles associated with TF.
Next, for the disease vocabulary, we adopted the Comparative Toxicogenomics Database’s (CTD)22 MEDIC23 resource. MEDIC is constructed by combining the Online Mendelian Inheritance in Man (OMIM), which provides comprehensive information on human genetic diseases, and terms in the disease categories offered by Medical Subject Headings (MeSH), which offer an efficient curation method for disease-related PubMed articles. As of March 2024, the MEDIC resource encompasses over 13,000 concepts and 78,000 synonyms, with regular updates to its vocabulary on a monthly basis. The extensive MEDIC vocabulary is widely utilized in biomedical algorithm research. For example, DNorm24, a tool specifically designed for disease normalization in clinical notes, utilizes MEDIC and has demonstrated exceptional performance during the 2013 ShARe/CLEF shared task. Also, MEDIC is effectively employed for disease Named Entity Recognition (NER) within PubTator Central25 which provides advanced automatic annotation tools for biomedical concepts, including genes and diseases, within both PubMed abstracts and PubMed Central full-text articles. Furthermore, MEDIC has been widely adopted in various studies that involve the construction of corpora based on PubMed abstracts22,26. Additionally, every disease term in MEDIC is assigned a corresponding MeSH Unique ID. This makes MEDIC an ideal disease vocabulary for creating a TFDR corpus, as it enables seamless integration with articles indexed in PubMed, ensuring precise and comprehensive disease term mapping. Therefore, in this study, CTD’s MEDIC was chosen as the disease vocabulary. However, MEDIC has certain limitations. Its polyhierarchical structure, where a disease may appear in multiple branches with varying descendants, can complicate disease categorization. Additionally, while MEDIC’s extensive coverage is beneficial, it may lack representation for rare or emerging diseases, and variations in terminology across different medical fields or regions could present further challenges.
Annotation workflow
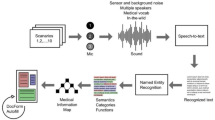
The workflow for constructing the TFDR corpus is depicted in Fig. 1. Initially, search queries were generated by combining the English expressions of TF listed in the traditional formula vocabulary, as explained in the preceding section. Subsequently, a total of 5,763 abstracts were downloaded from the PubMed database. For downloaded abstracts, TF mentions were automatically pre-annotated using a dictionary-based approach, while disease mentions were pre-annotated using the TaggerOne algorithm27. TaggerOne employs semi-Markov Models and also relies on the comprehensive MEDIC vocabulary, provided through the PubTator API. Out of the total 5,763 abstracts, 4,095 abstracts were identified to contain both TF mentions and disease mentions. Subsequently, a subset of 740 abstracts was randomly chosen for following annotation tasks. Although TaggerOne generally performs well, some limitations remain. Its reliance on a predefined lexicon can lead to missed terms, especially for rare or newly emerging terminology, and boundary inconsistencies within complex entity structures may affect normalization accuracy. As a result, some terms may have been overlooked during the initial identification. It should be noted that no manual validation was performed on the abstracts without identified mentions, leaving the possibility of undetected or misidentified entities. However, given TaggerOne’s robustness, the chance of significant omissions is considered low.
To construct the TFDR Corpus, annotators independently performed annotation tasks on TF mention, diseases mention, and key-sentences within PubMed abstracts. They also extracted relationships between TF and diseases mentions from the key-sentence, because key-sentence provides direct evidences to support the claimed relationships16. A total of six annotators participated in the construction of the TFDR corpus, all of whom were proficient in English and experts in traditional medicine, holding medical degrees in traditional medicine. Firstly, the annotators involved in the construction of the corpus received training on the objectives of corpus construction, annotation guidelines, and annotation tools. To ensure a comprehensive understanding of the annotation guidelines, a two-week training program was conducted. This training program involved utilizing a subset of 40 pre-annotated abstracts, carefully selected from the initial pool of 740 abstracts, to ensure effective training. These 40 abstracts were specifically chosen to cover a wide range of TF and disease terms, key-sentence selection, and different types of relations, allowing the application of the annotation guidelines during the training process. Through this training period, the annotators familiarized themselves with the annotation guidelines, ensuring a consistent and accurate annotation process. The team of annotators involved in the construction of the TFDR corpus comprised graduate students and clinical practitioners possessing specialized expertise in traditional medicine and diseases, ensuring the development of a corpus of high-quality. The annotators were divided into two groups, with three annotators independently performing annotations for each abstract. The annotation and curation process were conducted in four phases as outlined below:
Phase 1. Initial annotation
In this phase, each group of three annotators engaged in independent annotation tasks on approximately 40 pre-annotated abstracts per week. The annotation process for each group encompassed around 350 abstracts and spanned approximately 10 weeks. The assignment of annotators to their respective groups remained unchanged throughout the corpus construction process.
Phase 2. Review meeting
Following the completion of annotation tasks for the assigned abstracts each week, the annotators within each group convened regular meetings to discuss and address any discrepancies observed in the annotation results. These meetings aimed to reach a consensus and derive solutions for resolving any discrepancies or ambiguities identified during the annotation process.
Phase 3. Annotation revision
In this phase, the annotators conducted an independent round of annotation based on the solutions derived from the review meetings. However, the acceptance of these proposed solutions was determined by each annotator independently, taking into account their own judgment and discretion.
Phase 4. Final curation
The final annotation outcome for each abstract was derived by integrating the independent annotation results from the three annotators. In cases where discrepancies occurred between the two rounds of annotation, the majority vote principle was applied to establish the final annotation decision.
Annotation guidelines
The guidelines were developed to facilitate annotation workflows and ensure high inter-rater agreement for the creation of a high-quality TFDR corpus. The guidelines used in this study were drafted by analyzing those used in previous researches15,16,28 and adapting them to align with the objectives of this study. The guidelines were then updated through a series of tests, where they were applied to the annotation of article abstracts. After a two-week training program, a meeting was held with the annotators to make final revisions, addressing any gaps or inconsistencies in the guidelines. During the annotation and curation process, the guidelines were not modified further in order to maintain consistency throughout the corpus. The TFDR corpus workflow included four annotation steps: TF annotation, disease annotation, key-sentence annotation, and relation annotation within the key-sentence. To support an efficient annotation process, we employed WebAnno29, a web-based annotation tool designed for various linguistic annotations. The guidelines that annotators should refer to and follow in the annotation workflow are thoroughly documented in the accompanying guidelines, which can be accessed in the supplementary materials. In this section, we provide a summary of the main points as follows:
Annotation of traditional formula mention
-
Annotate the maximum span of traditional formula mentions. (e.g., “yukmi-jihwang-tang” rather than “jihwang-tang”)
-
Annotate all synonymous mentions. (e.g., Abbreviation definitions such as “Xiaochaihutang (XCHT)” are separated into two annotated mentions.)
-
Do not annotate product name or product number. (e.g., TJ-41 for the mention, Hocku-ekki-to (TJ-41))
-
Do not annotate substances or medicinal herbs comprising the traditional formula.
Annotation of disease mention
-
Annotate the most specific disease mentions with maximum span. (e.g., “Insulin-dependent diabetes mellitus” rather than “diabetes mellitus”.)
-
Annotate all synonymous mentions, including abbreviations possible to assume though not specified in the abstract.
-
Annotate each disease and symptom mentions separately, when the disease or symptom induced by disease. (e.g., “diabetes” and “cardiomyopathy” are annotated for the mention, “diabetes-induced cardiomyopathy”)
-
Annotate the mention representing the condition of disease (e.g., “severe dyspnea” rather than “dyspnea” and “dry cough” rather than “cough”)
-
Do not annotate mentions, if the prefix of it is “anti-”.
-
Do not annotate mentions concerning traditional medical diseases except “Yang Deficiency” and “Yin Deficiency”. (e.g., liver-wind stirring syndrome, fluid-retention syndrome, and so on are not annotated)
Annotation of key-sentence
-
Key-sentence is a condensed representation of the result or conclusion of the article. It should contain both traditional formula and disease mentions. It might contain multiple relations between them.
-
Annotate the title as the key-sentence, when no proper key-sentence is in the abstract.
-
Do not annotate the sentence as a key-sentence, if it merely refers to findings from previous studies.
Annotation of relations in the key-sentence
-
The Treatment of Disease relation is the “treatment”, “alleviation”, or “prevention” effects of the traditional formula on the disease. (e.g., “Using this model, Shakuyaku-kanzo-to was shown to relieve paclitaxel-induced painful peripheral neuropathy.” [PMID: 18472288])
-
The Cause of Side-effect relation is the “occurrence” or “exacerbation” effects of the traditional formula on the side-effect (e.g., “A case of pneumonitis induced by Bofu-tsusho-san” [PMID: 12692947])
-
The Association relation is annotated when, despite the co-occurrence of traditional formula and disease mentions in the key-sentence, the “description” or “correlation” between them is either unclear or not explicitly stated. (e.g., “Immunologic examination of Juzentaiho-to (TJ-48) in postoperative gastric cancer” [PMID: 2730049])
-
The Negative relation is the “ineffectiveness” of the traditional formula against the disease. (e.g., “However, given the high risk of bias among the trials, we could not conclude that YCWLP was beneficial to patients with hyperlipidemia.” [PMID: 27400466])
Based on the aforementioned guidelines, we present an annotated example abstract, as depicted in Fig. 2. In the example abstract, “Sishen Pill” and “SSP” are annotated as TF mention. Additionally, disease mentions which are “colitis”, “inflammatory bowel disease”, “IBD”, and “pathological damage to the colon” were annotated correctly. The concluding statement, which provides a concise summary of the entire abstract, was designated as a key-sentence and annotated accordingly. Within this sentence, TF mention “SSP” has Treatment of Disease relationship with disease mention “colitis.”
Disagreements
In the annotation results, we identified several instances of disagreement between annotators. According to our analysis, most of the disagreed cases occurred in the following situations:
-
Missed identification of TF abbreviations (e.g., “HLJDD” for “Huang-lian-Jie-du decoction”).
-
Differences in the scope of disease identification (e.g., “Cholestasis” vs. “Variable Cholestasis”).
-
Recurring discrepancies between annotators when identifying key sentences (e.g., “Our study showed YGW administration effectively alleviated BCAA metabolic disorder and improved gut dysbiosis” vs. “This study provides support for YGW administration with benefits for allergic asthma”).
-
Frequent disagreement in distinguishing between Treatment of Disease and Association when establishing the relationship between TF and diseases (e.g., in “Dachengqi decoction may promote the recovery of intestinal mucosal permeability and decrease the incidence of MODS and pancreatic infection in patients with severe acute pancreatitis,” annotators differed in identifying the relationship between Dachengqi decoction and pancreatic infection, as well as Dachengqi decoction and severe acute pancreatitis).
Annotation quality assessment
Due to the manual construction of the corpus by annotators utilizing their domain expertise and adhering to well-defined guidelines, the evaluation of corpus quality plays a vital role in assessing its comprehensiveness and utility. As mentioned previously, the annotation workflow of the TFDR corpus involved five steps. The inter-annotator agreement (IAA) was computed for each step to provide valuable insights into the reliability and consistency of TFDR corpus. As three annotators independently conducted the annotation tasks for constructing the TFDR corpus, the Fleiss’s kappa30 score was employed to compute the IAA scores. Fleiss’s kappa is a statistical measure utilized to evaluate the reliability of agreement among a predetermined number of evaluators when assigning categorical ratings or classifying items into multiple categories. In contrast to Cohen’s kappa, which is applicable to two raters, Fleiss’s kappa operates with an arbitrary number of raters. The computation of Fleiss’s kappa is performed using the following formula:
where \(\left(1-\bar{{P}_{e}}\right)\) factor represents the degree of agreement that can be attainable over chance, and \(\left(\bar{P}-\bar{{P}_{e}}\right)\) gives the degree of agreement that is actually achieved over chance. If the all annotators matched completely, then Fleiss’s kappa κ = 1. According to Viera et al.31, kappa values between 0.6 and 0.8 represent “substantial” agreement, while values above 0.8 can be interpreted as indicating “almost perfect” agreement. In this study, both strict and relaxed constraints were applied when calculating the IAA. The strict constraint was used when the annotations from the three annotators had the same entity type and identical offsets. On the other hand, the relaxed constraint was applied when the annotations from the three annotators shared the same entity type but had partially overlapping ranges.
Corpus evaluation
As an illustrative example of TFDR corpus’ validity, we conducted experiments within the realm of biomedical NLP, focusing on three key tasks: Named Entity Recognition (NER), Key-Sentence Recognition (KSR) and Relation Extraction (RE). NER involves the identification of words within unstructured text that correspond to predefined entities. KSR task evaluates whether a given sentence serves as a condensed representation of the result or conclusion of the article. Meanwhile RE pertains to the detection and classification of mentions representing semantic relationships within unstructured documents. Recent advancements in NLP, particularly with the rise of contextual language models based on the Transformer’s encoder architecture, have demonstrated exceptional performance, outperforming conventional machine learning methods. This is largely due to the Transformer encoder’s enhanced ability to capture contextual information bidirectionally, allowing it to better understand relationships between words and sentences, thus excelling in text and language comprehension. Notably, these advancements have translated into exceptional performance gains in NER, KSR and RE tasks as well. Firstly, BERT32 is composed solely of the encoder part of the Transformer architecture and employs Masked Language Model (MLM) and Next Sentence Prediction (NSP) during pre-training. These techniques enable BERT to learn bidirectionally, allowing it to capture context more effectively. BERT was pre-trained on a large corpus of general English text, including Wikipedia and BooksCorpus, and has demonstrated outstanding performance across various NLP tasks. Furthermore, fine-tuned BERT-based models have been developed for specific domains, such as SciBERT33, trained on computer science and biomedical domain papers. Another noteworthy model is BioBERT34, which initializes with BERT’s weights and further undergoes pre-training using additional corpora, specifically PubMed abstracts and PMC full-text articles.
While SciBERT and BioBERT are domain-specific models fine-tuned on BERT, ELECTRA35 and DeBERTa36 are language models that enhance BERT’s architecture and training methods to improve performance. ELECTRA has a structure similar to BERT but introduces an innovative training method. It consists of two networks: a generator and a discriminator. Instead of predicting tokens during training, it learns by distinguishing between original tokens and replaced tokens, leading to more efficient learning. As a result, ELECTRA can achieve similar or better performance than BERT with fewer resources. DeBERTa enhances the BERT and RoBERTa37 models through two innovative techniques. The first is the disentangled attention mechanism, where each word is represented by two separate vectors—one for its content and the other for its position. The second technique involves an improved mask decoder, which replaces the standard softmax output layer for predicting masked tokens during model pretraining. These advancements result in outstanding performance across a range of benchmarks.
Performance evaluation for the NER, KSR and RE downstream tasks on TFDR corpus utilized three metrics: micro-F1, macro-F1, and weighted-F1. The F1 score is defined as the harmonic mean of precision and recall. If all labels are of similar importance, the macro-F1 score is employed. For cases where importance is weighted towards labels with a greater number of samples, the weighted-F1 score is consulted. Additionally, when evaluating the overall model performance regardless of labels, the micro-F1 score is used.
Data Records
Corpus description
The TFDR corpus is available at Figshare38 and can also be accessed via its GitHub repository (https://github.com/KIOM-AIDoc/TFDR). Within the TFDR repository, the Codes folder contains Python scripts used for NER, KSR, and RE baseline experiments. The Corpus directory includes a ProcData subfolder, where the preprocessed datasets for these experiments are stored; each dataset is in a tab-separated text file format without column headings. Additionally, the RawData subfolder within the Corpus directory holds the original corpus files, formatted in an extended version of the PubTator corpus structure. This format includes three main sections: Text, NER, and RE information, organized as depicted in Fig. 3, with all data tab-separated. In the Text information segment, the paper’s title, abstract, and key-sentences are differentiated by the |t|, |a|, and |k| tags, respectively. The NER information encompasses details such as TF and disease entity offsets (e.g., 50 and 70), mentions (e.g., So-Cheong-Ryong-Tang), and entity types (e.g., Formula). Furthermore, the RE information presents data regarding the first entity (e.g., Start: 1431, End: 1435, Mention: SCRT) and the second entity (e.g., Start: 1490, End: 1535, Mention: chronic, perennial, and moderate to severe AR), along with the relation type between these entities (e.g., Treatment of Disease).
Corpus statistics
The TFDR is the first corpus annotated on TF and disease mentions, key-sentence, and relation within the key-sentence extracted from PubMed abstracts. The overall statistics of the TFDR corpus are presented in Table 1. Firstly, the corpus, generated from 740 PubMed abstracts, contains a total of 6,211 TF mentions and 7,166 disease mentions, with an average of 8.4 mentions and 9.7 mentions per abstract, respectively. Moreover, among the 744 key-sentences, 1,109 relations were annotated, indicating an average of 1.5 relationships between TF and diseases per key-sentence. Furthermore, as shown in Table 1, among the 1,109 relation annotations, the “Treatment of Disease” relationship accounts for the largest proportion at 83.3% (924), followed by “Association” at 12.8% (142). The “Cause of Side-effect” relationship represents 2.7% (30), while “Negative” stands at 1.2% (13).
Technical Validation
Inter-annotator agreement
In the process of building the TFDR corpus, two annotation phases are involved manual annotation by annotators: Initial annotation, which involved annotating pre-annotated abstracts, and annotation revision, which entailed re-annotating by reflecting the resolutions of review meeting. Therefore, IAA was measured for the outcomes of both annotation phases, and the results are presented in Table 2. In the Initial annotation phase, the IAA scores for strict constraints related to TF and disease mentions were 0.949 and 0.781, respectively. However, for the relaxed constraints, the IAA values showed improvement, reaching 0.958 for TF mentions and 0.821 for disease mentions. Notably, Table 2 illustrates that the IAA value for TF exceeded that of disease mentions. In terms of key-sentence annotation, the IAA was measured at 0.752, and for the annotation between TF and disease relations within key-sentences, the IAA was higher at 0.892. In the review meeting phase, annotators from each group engaged in discussions to address and resolve discrepancies in the initial annotation results. Through these discussions, they collectively derived solutions. The acceptance of these solutions was determined independently by the annotators, and they were subsequently incorporated during the annotation revision phase. The IAA scores for the annotation revision results, all approaching 0.99, indicate that the discussions among annotators during the review meeting phase were effective, leading to meaningful consensus and agreement.
Baseline evaluation
The goal of this evaluation wasn’t to achieve optimal performance, but rather to establish a baseline for future comparisons and demonstrate the usefulness of the TFDR corpus. In this study, we employed five distinct language models to fine-tune and evaluate their performance on the downstream tasks of NER, KSR and RE using TFDR corpus. The TFDR corpus was split into two distinct datasets: a training set consisting of 590 abstracts (79.7%) and a testing set containing 150 abstracts (20.3%), making a total of 740 abstracts. These abstracts were randomly assigned to the training and testing sets to ensure that the model was trained on a broad and diverse sample of data while keeping a separate set of documents unseen during training for unbiased evaluation. For the NER task, language models determined entity boundaries and labels based on the probabilities generated by a classifier for each token in the input sentence. The KSR task assesses whether a given sentence is key-sentence of abstract or not. To avoid generating skewed training and testing data due to an excess of non-key-sentences, non-key-sentences were randomly selected to maintain a ratio of 1:5 between key- and non-key-sentences. Subsequently, in the RE downstream task, the labels representing relationships between TF and diseases present in the input sentences were inferred by a trained classifier. To mitigate bias towards specific TF and disease tokens, entities existing within the input sentences were replaced with “@FORMULA@” and “@DISEASE@” placeholders during the pre-processing.
The performance evaluation of NER employed a strict matching methodology, assessing the precise alignment of predicted entity types and boundaries by the classifier. The analysis of the NER downstream task results, as shown in Table 3, indicates that DeBERTa outperformed the other models, achieving the highest micro-F1 (89.96.%), macro-F1 (89.71%), and weighted-F1 (89.84%) scores, demonstrating its superior ability to generalize across different entities. BioBERT followed closely with strong scores across all metrics, particularly in macro-F1 (90.14%), indicating its effectiveness in the biomedical domain. SciBERT and ELECTRA performed similarly, with ELECTRA slightly outperforming SciBERT in all three metrics. BERT had the lowest performance among the models but still demonstrated solid NER capabilities. Overall, DeBERTa and BioBERT showed the best performance in this task.
The analysis of the KSR task results shows that DeBERTa and BioBERT outperformed the other models, achieving the highest scores across all metrics. These results can be confirmed in Table 4. DeBERTa achieved the highest macro-F1 score (86.98%), indicating its strong ability to handle class imbalances, while both DeBERTa and BioBERT shared the top micro-F1 score (93.20%), reflecting their overall accuracy. ELECTRA followed closely, with solid performance across all metrics, outperforming SciBERT and BERT, which had slightly lower macro-F1 scores. Overall, DeBERTa and BioBERT demonstrated the best generalization capabilities making them the most effective models for the KSR task.
The analysis of the RE results, as shown in Table 5, indicates that DeBERTa achieved the highest macro-F1 score (54.02%), along with a strong weighted-F1 score (80.90%). ELECTRA closely followed with comparable performance, matching BioBERT in micro-F1 (83.93%) while slightly outperforming it in macro-F1 and weighted-F1. SciBERT also performed well, achieving solid scores across all metrics. BERT had the lowest performance, particularly in macro-F1 (51.74%), indicating some challenges with class imbalance, though its micro-F1 and weighted-F1 scores remained competitive. Overall, DeBERTa and ELECTRA emerged as the top models for RE tasks, with DeBERTa excelling in handling diverse relation types compared to the other models.
In comparison to the “Treatment of Disease” relationship in the TFDR corpus, the remaining three types of relationships appear to be relatively scarce. This scarcity is reflected in lower macro-F1 scores compared to micro/weighted-F1 scores in RE downstream task. When training language models on highly imbalanced data, the lower macro-F1 score compared to the micro/weighted-F1 score highlights the challenges in handling class imbalance. This imbalance causes the model to be biased toward the majority class, leading to high performance on classes with more instances (reflected in the micro-F1) while underperforming on minority classes, which significantly lowers the macro-F1 score. The model struggles to learn meaningful representations for the minority classes due to insufficient data, leading to poor recall and precision for those classes. To address this, techniques such as class weighting, data augmentation, over-sampling of minority classes, or under-sampling of majority classes can be employed. Additionally, using focal loss or synthetic data generation methods like SMOTE39 can help mitigate the impact of class imbalance and improve the model’s performance across all classes.
Case study
From the 4,095 pre-annotated abstracts, 740 abstracts were randomly selected and manually annotated to construct the TFDR corpus. To ensure the unbiased sampling, we conducted an analysis of TF and disease distribution. The results confirmed no significant differences between the 740 selected abstracts and the remaining 3,355 unselected abstracts. However, the unselected group contained TF and disease terms that did not appear in the TFDR corpus. To analyze how the NER, KSR, and RE models developed in the study would perform on papers containing unseen TF and disease terms, we conducted a case study on 10 papers that met these criteria. The models successfully conducted NER, KSR, and RE downstream tasks for these 10 papers. Here are some notable cases:
-
TF recognition: The model accurately detected the term Guifu Dihuang Pill and its abbreviation GFDHP. However, in a different sentence, it mistakenly recognized G and D as separate abbreviations for TF [PMID: 34777538].
-
Disease recognition: While TaggerOne incorrectly recognized only arthritis as the disease in gouty arthritis, the NER model correctly identified the full term [PMID: 33505502]. However, the model failed to recognize mental fatigue [PMID: 34457018].
-
Key-sentence recognition: The sentence “Chaige Jieji Decoction recorded in the Six Books of Exogenous Febrile Disease could be used to treat exterior syndrome due to wind-cold and heat caused by stagnation” was incorrectly identified as a key sentence [PMID: 31872718].
-
Relation extraction: It was interesting that the model identified the relationship between Chaige Jieji Decoction and Exogenous Febrile Disease as an “Association” in the above sentence. In another sentence, “Chaige Jieji Decoction can not only treat exogenous diseases but also treat nosocomial infections in critically ill patients during hospitalization, “the model correctly predicted the relationship between Chaige Jieji Decoction and exogenous diseases and nosocomial infections as a “Treatment of Disease”, while critically ill was correctly linked with an “Association” relation [PMID: 31872718].
Building upon TFDR corpus
In this study, we have developed a high-quality corpus focused on the relationship between TF and diseases, named TFDR, and provided experimental validation of its effectiveness for tasks such as NER, KSR, and RE in the context of TCM and biomedical domain. To the best of our knowledge, the TFDR corpus is the first of its kind. By making this corpus freely available, we aim to facilitate the development of supervised TF-disease relation models and stimulate the creation of related applications. The potential downstream applications of this corpus are vast: a high-performing model trained on TFDR could be integrated into Clinical Decision Support Systems (CDSS) to assist healthcare professionals in recommending TFs based on a patient’s symptoms or diseases40. Additionally, it could aid drug discovery and repurposing by identifying TF-disease relationships41, and help build comprehensive knowledge graphs that encapsulate traditional medical knowledge from TF-disease interactions42. Given these possibilities, we believe that the tasks supported by TFDR are well-suited for novel shared tasks within the research community. However, it is important to note that the TFDR corpus was created using the TF vocabulary from the traditional Korean medicine ontology, which may make it more specialized for Korean TFs or older TFs found in ancient medical texts. Therefore, researchers and developers should consider this limitation when using TFDR for future applications.
Usage Notes
TFDR corpus is made available under the Creative Commons Attribution 4.0 International Public License (CC-BY). The baseline evaluation code is available at https://github.com/KIOM-AIDoc/TFDR/tree/main/Codes.
Code availability
The Python codes for the NER, KSR, and RE evaluation presented in Technical Validation can be accessed from https://doi.org/10.6084/m9.figshare.2707367238 and https://github.com/KIOM-AIDoc/TFDR.
References
Luo, J., Wu, M., Gopukumar, D. & Zhao, Y. Big data application in biomedical research and health care: A literature review. Biomed. Inf. Insights. 8, BII.S31559 (2016).
Liu, Z. et al. AI-based language models powering drug discovery and development. Drug Discovery Today. 26, 2593–2607 (2021).
Yang, Y. et al. A survey of information extraction based on deep learning. Appl. Sci. 12, 9691 (2022).
Cho, H., Kim, B., Choi, W., Lee, D. & Lee, H. Plant phenotype relationship corpus for biomedical relationships between plants and phenotypes. Sci. Data. 9, 235 (2022).
Rizvi, S. A. A., Einstein, G. P., Tulp, O. L., Sainvil, F. & Branly, R. Introduction to traditional medicine and their role in prevention and treatment of emerging and re-emerging diseases. Biomol. 12, 1442 (2022).
Jia, W. et al. The rediscovery of ancient Chinese herbal formulas. Phytother. Res. 18(8), 681–686 (2004).
Kim, J. H. Current status of use of traditional medicine and complementary alternative medicine in worldwide. Policy Report of Health Insurance Review & Assessment Service. July, 44–48 (2008).
Nguyen, M. N. T. & Ho-Huynh, T. D. Selective cytotoxicity of a Vietnamese traditional formula, Nam Dia long, against MCF-7 cells by synergistic effects. BMC Complement. Altern. Med. 16, 1–10 (2016).
Liu, C. & Huang, Y. Chinese herbal medicine on cardiovascular diseases and the mechanisms of action. Front. Pharmacol. 7, 469 (2016).
Zhou, X. et al. Inhibition activity of a traditional Chinese herbal formula Huang-Lian-Jie-Du-Tang and its major components found in its plasma profile on neuraminidase-1. Sci. Rep. 7(1), 15549 (2017).
Pirintsos, S. et al. From traditional ethnopharmacology to modern natural drug discovery: A methodology discussion and specific examples. Mol. 27, 4060 (2022).
Bunescu, R. et al. Comparative experiments on learning information extractors for proteins and their interactions. Artif. Intell. Med. 33, 139–155 (2005).
Pyysalo, S. et al. Bioinfer: a corpus for information extraction in the biomedical domain. BMC bioinf. 8, 1–24 (2007).
Hamon, T., Tabanou, V., Mougin, F., Grabar, N. & Thiessard, F. POMELO: Medline corpus with manually annotated food-drug interactions. BiomedicalNLP@RANLP. 73–80 (2017).
Kim, B., Choi, W. & Lee, H. A corpus of plant-disease relations in the biomedical domain. PLoS One. 14, e0221582 (2019).
Kim, S., Choi, Y., Won, J. H., Oh, J. M. & Lee, H. An annotated corpus from biomedical articles to construct a drug-food interaction database. J. Biomed. Inf. 126, 103985 (2022).
Zhang, T., Wang, Y., Wang, X., Yang, Y. & Ye, Y. Constructing fine-grained entity recognition corpora based on clinical records of traditional Chinese medicine. BMC Med. Inf. Decis. Making. 20, 1–17 (2020).
Sun, Y. et al. Leveraging a joint learning model to extract mixture symptom mentions from traditional Chinese medicine clinical notes. Biomed Res. Int. 2022, 2146236 (2022).
Yang, X., Wu, C., Nenadic, G., Wang, W. & Lu, K. Mining a stroke knowledge graph from literature. BMC bioinf. 22, 1–19 (2021).
Vaswani, A., et al. Attention is all you need. Adv. Neur. In. 30, (2017).
Kim, S. K. et al. Models and representations of formulas in Korean medicine information systems. J Korean Med. 35, 41–49 (2014).
Davis, A. P. et al. Comparative Toxicogenomics Database (CTD): update 2023. Nucleic Acids Res. 51(51), 1257–1262 (2022).
Davis, A. P., Wiegers, T. C., Rosenstein, M. C. & Mattingly, C. J. MEDIC: a practical disease vocabulary used at the comparative toxicogenomics database. Database. 2012, bar065 (2012).
Robert, L., Rezarta, I. D. & Zhiyong, L. U. DNorm: disease name normalization with pairwise learning to rank. Bioinf. 29, 2909–2917 (2013).
Wei, C. H., Allot, A., Leaman, R. & Lu, Z. PubTator central: automated concept annotation for biomedical full text articles. Nucleic Acids Res. 47, W587–W593 (2019).
https://ctdbase.org/about/publications/ Citing CTD/Publications/Use (2024).
Robert, L. & Zhiyong, L. U. TaggerOne: joint named entity recognition and normalization with semi-Markov models. Bioinf. 32, 2839–2846 (2016).
Dog˘an, R. I., Leaman, R. & Lu, Z. NCBI disease corpus: A resource for disease name recognition and concept normalization. J. Biomed. Inform. 47, 1–10 (2014).
Eckart de Castilho, R., et al. A Web-based tool for the integrated annotation of semantic and syntactic structures. In Proceedings of the LT4DH workshop at COLING 2016, Osaka, Japan, (2016).
Fleiss, J. L. Measuring nominal scale agreement among many raters. Psychol. bull. 76, 378–382 (1971).
Viera, A. J. & Garrett, J. M. Understanding interobserver agreement: the kappa statistic. Fam. Med. 37, 360–363 (2005).
Devlin, J., Chang, M. W., Lee, K. & Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. In Proceeding of the NAACL-HLT 2019, Minneapolis, USA, (2019).
Beltagy, I., Lo, K. & Cohan, A. Scibert: A pretrained language model for scientific text. arXiv preprint arXiv:1903.10676 (2019).
Lee, J. et al. BioBERT: a pretrained biomedical language representation model for biomedical text mining. Bioinf. 36, 1234–1240 (2019).
Clark, K. Electra: Pre-training text encoders as discriminators rather than generators. arXiv preprint arXiv:2003.10555 (2020).
He, P., Liu, X., Gao, J., & Chen, W. Deberta: Decoding-enhanced bert with disentangled attention. arXiv preprint arXiv:2006.03654 (2020).
Liu, Y. Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv:1907.11692 (2019).
Yea, S. J., Jang, H., Kim, S. Y., Lee, S. H. & Kim, J. U. TFDR corpus: annotated corpus for traditional formula-disease relationships in biomedical articles. Figshare. https://doi.org/10.6084/m9.figshare.27073672 (2024).
Chawla, N. V., Bowyer, K. W., Hall, L. O. & Kegelmeyer, W. P. SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. 16, 321–357 (2002).
Geng, W. et al. Model-based reasoning of clinical diagnosis in integrative medicine: real-world methodological study of electronic medical records and natural language processing methods. JMIR Med. Inform. 8(12), e23082 (2020).
Zhang, Y. et al. A Core Drug Discovery Framework from Large‐Scale Literature for Cold Pathogenic Disease Treatment in Traditional Chinese Medicine. J. Healthc. Eng. 2021(1), 9930543 (2021).
Gao, R. & Li, C. Knowledge question-answering system based on knowledge graph of traditional Chinese medicine. Proceedings of IEEE 9th Joint International Information Technology and Artificial Intelligence Conference (ITAIC). 9, 27–31 (2020).
Acknowledgements
This research was supported by AI Traditional Korean Medical Doctor Project (KSN1824130, KSN1923111) of Korea Institute of Oriental Medicine.
Author information
Authors and Affiliations
Contributions
S.Y. designed the study, supervised the annotation campaign, evaluated IAA and baseline performance and wrote the manuscript. H.J. designed the study, supported the annotation campaign, evaluated baseline performance and wrote the manuscript. S.K. designed the study and wrote the manuscript. S.L. initiated the design of the study and advised the writing of the manuscript. J.K. advised the design of the study and the writing of the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yea, S., Jang, H., Kim, S. et al. Annotated corpus for traditional formula-disease relationships in biomedical articles. Sci Data 12, 26 (2025). https://doi.org/10.1038/s41597-025-04377-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04377-2