Abstract
The trabecular meshwork within the outflow apparatus is critical in maintaining intraocular pressure homeostasis. In vitro studies employing primary cell cultures of the human trabecular meshwork (hTM) have conventionally served as surrogates for investigating the pathobiology of TM dysfunction. Despite its abundant use, translation of outcomes from in vitro studies to ex vivo and/or in vivo studies remains a challenge. Given the cell heterogeneity, performing single-cell RNA sequencing comparing primary hTM cell cultures to hTM tissue may provide important insights on cellular identity and translatability, as such an approach has not been reported before. In this study, we assembled a total of 14 primary hTM in vitro samples across passages 1–4, including 4 samples from individuals diagnosed with glaucoma. This dataset offers a comprehensive transcriptomic resource of primary hTM in vitro scRNA-seq data to study global changes in gene expression in comparison to cells in tissue in situ. We have performed extensive preprocessing and quality control, allowing the research community to access and utilize this public resource.
Similar content being viewed by others
Background & Summary
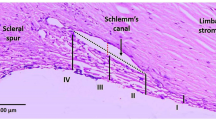
Primary open angle glaucoma (POAG) is a devastating ocular disorder resulting in irreversible vision loss. While the precise etiology and progression of POAG are complex, reducing intraocular pressure (IOP) is the only modifiable risk factor for managing visual field loss. The trabecular meshwork (TM) in the anterior segment of the eye is a major site of egress for aqueous humor1. Dysfunction of the TM, contributed by tissue resident cells and the extracellular matrix (ECM), is thought to result in increased resistance to aqueous drainage and ocular hypertension (OHT)2,3. The TM is thought to be anatomically heterogeneous and is comprised of 3 regions: the juxtacanalicular region (JCT) within the cribriform region, corneoscleral meshwork, and uveoscleral meshwork, of which the area around the juxtacanalicular region (JCT) of the meshwork and the inner wall cells of the Schlemm’s canal is considered the major site of resistance to outflow4,5,6,7,8. The primary intervention to reducing OHT is achieved through targeting either reduction of aqueous production (e.g. α-adrenergic agonists, β-blockers, carbonic anhydrase inhibitor), or via increasing outflow via the unconventional pathway (e.g. prostaglandins, miotic & cholinergic agents). More recently, Rho kinase inhibitors and nitric oxide donor drugs that target the conventional outflow apparatus have been approved for use.
Understanding the molecular pharmacology and mechanism of action of drugs has often relied on primary TM cells cultured in vitro prior to conduction of ex vivo or in vivo studies. However, translation of in vitro efficacy to pre-clinical and subsequently clinical outcomes remains a challenge. Differences in tissue anatomy of ocular structures, physiology, microenvironment, and species all contribute to the inability to replicate the complexities of an organism in vitro. The physiology and anatomy of the TM is complex with functional heterogeneity observed in the form of segmental regions of high, intermediate and low flow9. Segmental heterogeneity is associated with changes in extracellular matrix composition, biomechanical properties, and molecular signaling pathways in cells10,11,12,13,14. To further understand this complex tissue, single cell transcriptomic studies demonstrate heterogeneity in cell types within the conventional pathway with 12-to-19 distinct cell types identified with region-specific expression of candidate genes to define cellular identity15,16. In contrast, in vitro cell culture of primary human TM (hTM) cells originate with isolation of these cells from TM tissue dissected out of human donor anterior segment, corneal rims or whole globes. Though these primary cells are utilized for in vitro studies in early passages (∼2–6), there is a growing recognition among the community that cellular identity is heterogeneous, may change with time, and is variable from donor-to-donor. Furthermore, there is a considerable interest in the concept of mechanical memory of cells that may help define cellular identity and phenotypic characterizations.
In this study, primary hTM cells were isolated and cultured from non-glaucomatous and glaucomatous donors following tissue dissection, validated through cobblestone morphologic appearance and dexamethasone-induced myocilin, and compared with freshly dissected human TM tissue via single cell transcriptomics. Several TM cell specific marker genes were identified, e.g. Chitinase 3 Like 1 (CHI3L1), matrix gla protein (MGP), and myocilin MYOC albeit within different clusters. Prior comprehensive transcriptomic analysis demonstrates the aforementioned genes to be present in the TM and other ocular tissues15,16,17,18 affirming that the cells characterized are indeed from the appropriate tissue isolated (see Fig. 1 for workflow). When the transcriptome of these primary hTM cells were superimposed with those of TM tissue, a striking divergence in cell composition was observed. Specifically, we observe dramatically reduced cell heterogeneity and changes between transcriptomics profiles in the in vitro culture compared to in vivo tissue.
Methods
Isolation of donor primary hTM cells
Primary hTM cells were isolated from human donor corneal rims deemed unsuitable for transplantation, and characterized as described previously19,20. All donor tissues for primary cell culture were procured from Saving Sight eye bank (Kansas city, MO). All primary cell culture donor cells (Tables 1, 2) used in this study were isolated (between 2012 and 2022), validated, stored in liquid nitrogen, or used in prior studies from our lab14,21,22,23,24. Frozen cells were thawed with 15 mL pre-warmed media (37 °C, DMEM:F12 = 1:1, with 20% FBS, 1% Penn/Step/fungizone) and centrifuged at 300 g, 4 °C for 5 minutes. Cells were washed with media again and suspended in 0.04% BFA. Cell viability was checked with trypan blue.
Donor tissues
TM tissue for scRNAseq was dissected from donor anterior segments/whole globes obtained from Lions Eye Bank (Baylor College of Medicine, Houston, TX) within 4–6 hours post-mortem. Dissection was performed in accordance with consensus guidelines19.
Single-cell RNA sequencing
Resuspended single cells were loaded on a 10X Chromium controller for obtaining single cell Gel Beads-In-Emulsions. scRNA-seq libraries were generated using 10X Chromium Single Cell 3′ reagent kits v3.1 (10X Genomics) following the manufacturer’s recommendations (https://www.10xgenomics.com). Sequencing was performed on Illumina Novaseq. 6000 (http://www.illumina.com) at the Single Cell Genomics Core at Baylor College of Medicine.
Meta-analysis of scRNA-seq datasets
Preprocessing of scRNA-seq Datasets
Raw sequencing reads were processed using the CellRanger v6.1.2 (10X Genomics) pipeline, against the hg38 reference genome (https://cf.10xgenomics.com/supp/cell-exp/refdata-gex-GRCh38-2020-A.tar.gz). Then, quality control was performed separately for each sample through a quality control pipeline (https://github.com/lijinbio/cellqc)25,26. Real cells were filtered by dropkick v1.2.827 and ambient RNA was removed using SoupX v1.6.228. Doublets were detected and removed using DoubletFinder29. After merging all samples, cells were further filtered manually (features >300, transcript (UMI) counts >500, mitochondrial percentage <5%) (Table 3).
Data integration and clustering
The raw counts from each sample were merged, and the scVI model (n_layers = 2, n_latent = 30) from scvi-tools v0.19.530 was used to infer a latent space of 30 dimensions from the raw counts of the top 2000 highly variable genes (HVGs) calculated by scanpy v1.9.131. Sample ID was provided as a batch covariate for the scVI model. The latent space was further reduced using UMAP (min_dist = 0.3), and leiden clustering (resolution = 0.5) was performed on the scVI latent space, using a k-nearest neighbor graph (neighbors = 20).
Cell clustering and cell type annotation
Cell cycle effects were removed using Seurat32. The clustering resolution was validated using sccaf33. Initial cell type annotation was performed using scPred34 with a reference dataset generated from in vivo tissue samples from our lab. Clusters were re-annotated using known cell type marker genes and cell types were assigned to each cluster using scanpy31 with specific markers from previous publications15,16. The detailed markers for each cell type are listed in Table 4. Cell proportion plots were generated using dittoseq35.
Differentially expressed gene analysis
To identify genes that are differentially expressed between cell types, genes specifically expressed in each cluster were identified and the top 5 genes expressed in each cluster were ranked using Seurat32.
Data integration with in vivo dataset
Cell type of three samples of in vivo hTM cells was determined with scPred individually and then an integrated object of only in vivo data was compared with the same known cell type marker genes to determine validity of the in vivo samples. Data integration with the tissue culture data was then performed following the same parameters and protocol as above. Then, the combined tissue + culture object was once again compared with the known cell type marker genes and a disparity in gene expression was observed.
Data Records
Raw reads of all samples and processed data files including integrated data were deposited in the Gene Expression Omnibus (GEO, https://ncbi.nlm.nih.gov/geo) of the National Center for Biotechnology Information (NCBI) as FASTQ files with accession number GSE26323036.
Technical Validation
Prior to sequencing experiments, hTM cells were simultaneously validated by documenting Myoc mRNA expression in response to 100 nM dexamethasone treatment for 3 days (Fig. 2). Quality control was performed on each sample independently with an average of 6000 cells profiled, yielding a total of about 82k cells (Table 3). There are an average of 2300 gene counts and 8400 UMI counts over the 14 samples (Fig. 3(a)). After integration, we affirmed that batch effects were not significant (Fig. 3(b)).
By performing clustering analysis of all the cells, a total of 5 clusters were obtained. Of the 5 clusters identified, 3 clusters (TM_Culture_Cell, Proliferating_Culture_Cell, and Stressed_Culture_Cell) are shared by all donors (Fig. 4(a)). The other two clusters are only present in the hTM_7987 sample (Fig. 4(b)). This suggests that the population of hTM cells in culture is non-uniform and exhibits some heterogeneity. Several TM cell specific marker genes were identified; e.g. CHI3L1, MGP, and MYOC within different clusters. While MYOC was primarily expressed in the TM_Culture_Cell cluster, CHI3L1 and MGP were abundant in the BeamCell cluster. When overlaid with cell-specific markers previously reported, PDPN, CEMIP and CHI3L1 were abundant in the BeamCell cluster, while MYOC remained enriched in the TM_Culture_Cell cluster. Matrix proteins, collagens 1/6, FBLN, FN, POSTN, and DCN were enriched in clusters BeamCell and Fibroblast, while PCOLCE was enriched in clusters TM_Culture_Cell and Proliferating_Culture_Cell (Fig. 4(c)). Prior comprehensive transcriptomic analysis demonstrates the aforementioned genes to be present in the TM and other ocular tissues15,16,17,18 affirming that the cells characterized are indeed from the appropriate tissue isolated. Further, pairwise correlation analysis of psuedo-bulk transcriptomic profile expression of cells was made comparing our study with the previous study16 (Supplementary Fig. 1). Data demonstrated significant correlation, indicating good concordance in cell identities and characterization between both studies.
With the tissue data, we used the same QC pipeline with the same parameters for prepossessing. In the tissue samples alone, there are an average of 8100 cells profiled, yielding a total of about 24k cells with an average of 2300 gene counts and 6400 UMI counts. We assigned cell types using scPred to yield 9 defined cell types with a few unassigned cells (Fig. 5(a)). We validated the clustering and cell type assignment with previously identified marker genes15,16,17,18 and confirmed that the data were suitable for further analysis.
Usage Notes
Our dataset will be useful for a variety of studies pertaining to understanding the identity of primary hTM cells, including studies to translate pre-clinical in vitro models to in vivo models mimicking disease, relevant biophysical and biochemical cues mimicking the native cellular microenvironment, and choice and type of in vitro models utilized for investigations. Here we provide a comparison between our dataset and tissue data generated by our lab as a usage example:
When the transcriptome of these primary hTM cells were superimposed with those of TM tissue, a striking divergence in cell identity was observed. Specifically, reduced cell heterogeneity and dramatic changes of transcriptomics profiles in the in vitro culture compared to in vivo tissue (Fig. 6). Importantly, we note that a comparison of primary hTM cells cultured from glaucomatous and non-glaucomatous donors demonstrated no significant differences in cell type markers or clusters identified (Supplementary Fig. 2,3). Furthermore, regardless of disease state, primary hTM cells differ in their transcriptome profile compared with human tissue. We also note that primarily the tissues and cells were isolated from donors with similar reported ancestry (white). As such, differences observed between cultured cells and tissues is unlikely due to race. It is important to note that the primary hTM cells characterized in this study were previously frozen down in liquid N2 and are from early passages (up to 2, or as mentioned in Table 1). It is unclear why and at what stage these putative markers may be lost in culture. We are also uncertain whether, at any point during culture, these cells may have de-differentiated from a specific population of proliferating cells, as the heterogeneity observed in vitro is significantly reduced compared to that in the tissue. Nevertheless, while it is currently out of scope, we anticipate that future investigations will perform secondary validation efforts comparing cell specific marker expression in both primary TM cells in culture and tissues.
Data integration for comparison of in vivo and in vitro data. (a) UMAP by sample type with separation of clusters between primary cell culture and post-mortem tissue. (b) UMAP by cell type with separation of clusters between primary cell culture and post-mortem tissue. (c) Dot Plot showing marker gene expression by cluster between both sample types.
Our group and others have consistently reported that biophysical and biochemical cues from the cell culture microenvironment (topography, stiffness, ECM coating, stretch, 2D vs 3D) have profound impact on transcriptomic, proteomic, signaling pathways, and response to drugs in vitro37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56. Thus it is feasible to infer that while mechanical cues may drive hTM cell function as a function of substrate properties, the initial culture conditions in which these cells were first isolated and expanded may have also profound impact on selection of cells for propagation, proliferation, and (de-)differentiation. That mechanical memory and plasticity exists in maintaining cell identity through epigenetic regulation has been previously postulated57,58,59,60,61,62,63,64,65. However, whether such a phenomenon exists in primary hTM cells remains to be further determined. Emerging evidence suggests chromatin remodeling implicating significant changes to cellular and epigenetic plasticity may significantly alter cell fate with prolonged exposure to rigid culture environments such as tissue culture plastic62,63,66,67,68. Newer studies have also suggested that the cytoskeleton and transcriptional elements may have additional roles in cellular plasticity and memory, though the evidence for such is currently limited69,70,71. It is further interesting to note that cellular memory to external and innate stimuli is well documented for immune cells. However, whether such responses translate to non-immune cells is unclear although it is likely such mechanisms may be conserved. Whether these responses translate in the form of ECM remodeling, transcriptional & translational regulation, cytoskeletal reorganization, cell division and/or phenotypic outcomes warrants mechanistic investigation. For example, the evidence pertaining DNA methylation/cellular differentiation is shown to be dependent on cell type (e.g. stem cells vs terminally differentiated cells72,73,74) and substrate mechanical properties. The above studies were done in vitro and lacked a direct comparison to cells in vivo consequently highlighting some paucity in our understanding of these phenomena. Since all primary hTM cells are are terminally differentiated cells that are primarily cultured and expanded on rigid plastic substrates, it is only natural to infer that the process of divergence in cell identity as observed in this study likely starts immediately after initial isolation. A systematic study is critically needed to confirm this, and this may subsequently allow for the development of appropriate culture conditions and microenvironments which better maintain primary TM cell identity. Since a mechanistic approach was not taken in this study to ascertain why, how and when the divergence in cell identity comparing primary culture to tissues occurs, we point the audience to relatively recent manuscripts/editorials that attempt to draw highlight to this75,76,77. With increasing efforts in the use of organoids, microphysiological systems, bioprinting etc, integration of physiological, chemical, mechanical, spatial profiling, and multi-cell complexities may help develop better models for studying outflow homeostasis and dysfunction75,78,79.
All raw RNA sequencing data are stored in FASTQ files, and processed.h5ad files are also available for use.
Code availability
The source code, including code to generate all figures, has been uploaded to GitHub: https://github.com/RCHENLAB/TM_culture_manuscript.
References
Stamer, W. D. & Acott, T. S. Current understanding of conventional outflow dysfunction in glaucoma. Curr. Opin. Ophthalmol. 23, 135–143, https://doi.org/10.1097/icu.0b013e32834ff23e (2012).
Acott, T. S. & Kelley, M. J. Extracellular matrix in the trabecular meshwork. Exp. Eye Res. 86, 543–561, https://doi.org/10.1016/j.exer.2008.01.013 (2008).
Vranka, J. A., Kelley, M. J., Acott, T. S. & Keller, K. E. Extracellular matrix in the trabecular meshwork: Intraocular pressure regulation and dysregulation in glaucoma. Exp. Eye Res. 133, 112–125, https://doi.org/10.1016/j.exer.2014.07.014 (2015).
Johnson, M., Shapiro, A., Ethier, C. R. & Kamm, R. D. Modulation of outflow resistance by the pores of the inner wall endothelium. Investig. ophthalmology I& visual science 33, 1670–1675 (1992).
Tamm, E. R. Functional Morphology of the Trabecular Meshwork Outflow Pathways, 40–46 (Elsevier, 2015).
Tamm, E. Functional Morphology of the Trabecular Meshwork*, 157–163 (Elsevier, 2010).
Tamm, E. R. The trabecular meshwork outflow pathways: Structural and functional aspects. Exp. Eye Res. 88, 648–655, https://doi.org/10.1016/j.exer.2009.02.007 (2009).
Tamm, E. R. & Fuchshofer, R. What increases outflow resistance in primary open-angle glaucoma? Surv. Ophthalmol. 52, S101–S104, https://doi.org/10.1016/j.survophthal.2007.08.002 (2007).
de Kater, A. W. Patterns of aqueous humor outflow in glaucomatous and nonglaucomatous human eyes: A tracer study using cationized ferritin. Arch. Ophthalmol. 107, 572, https://doi.org/10.1001/archopht.1989.01070010586035 (1989).
Vranka, J. A., Bradley, J. M., Yang, Y.-F., Keller, K. E. & Acott, T. S. Mapping molecular differences and extracellular matrix gene expression in segmental outflow pathways of the human ocular trabecular meshwork. PLOS ONE 10, e0122483, https://doi.org/10.1371/journal.pone.0122483 (2015).
Cha, E. D., Xu, J., Gong, L. & Gong, H. Variations in active outflow along the trabecular outflow pathway. Exp. Eye Res. 146, 354–360, https://doi.org/10.1016/j.exer.2016.01.008 (2016).
Vranka, J. A. & Acott, T. S. Pressure-induced expression changes in segmental flow regions of the human trabecular meshwork. Exp. Eye Res. 158, 67–72, https://doi.org/10.1016/j.exer.2016.06.009 (2017).
Vranka, J. A. et al. Biomechanical rigidity and quantitative proteomics analysis of segmental regions of the trabecular meshwork at physiologic and elevated pressures. Investig. Opthalmology I& Vis. Sci. 59, 246, https://doi.org/10.1167/iovs.17-22759 (2018).
Dhamodaran, K. et al. Endogenous expression of notch pathway molecules in human trabecular meshwork cells. Exp. Eye Res. 216, 108935, https://doi.org/10.1016/j.exer.2022.108935 (2022).
Patel, G. et al. Molecular taxonomy of human ocular outflow tissues defined by single-cell transcriptomics. Proc. Natl. Acad. Sci. 117, 12856–12867, https://doi.org/10.1073/pnas.2001896117 (2020).
van Zyl, T. et al. Cell atlas of aqueous humor outflow pathways in eyes of humans and four model species provides insight into glaucoma pathogenesis. Proc. Natl. Acad. Sci. 117, 10339–10349, https://doi.org/10.1073/pnas.2001250117 (2020).
Carnes, M. U., Allingham, R. R., Ashley-Koch, A. & Hauser, M. A. Transcriptome analysis of adult and fetal trabecular meshwork, cornea, and ciliary body tissues by rna sequencing. Exp. Eye Res. 167, 91–99, https://doi.org/10.1016/j.exer.2016.11.021 (2018).
Wolf, J. et al. The human eye transcriptome atlas: A searchable comparative transcriptome database for healthy and diseased human eye tissue. Genomics 114, 110286, https://doi.org/10.1016/j.ygeno.2022.110286 (2022).
Keller, K. E. et al. Consensus recommendations for trabecular meshwork cell isolation, characterization and culture. Exp. Eye Res. 171, 164–173, https://doi.org/10.1016/j.exer.2018.03.001 (2018).
Morgan, J. T. et al. Human trabecular meshwork cells exhibit several characteristics of, but are distinct from, adipose-derived mesenchymal stem cells. J. Ocular Pharmacol. Ther. 30, 254–266, https://doi.org/10.1089/jop.2013.0175 (2014).
Raghunathan, V. K. et al. Glaucomatous cell derived matrices differentially modulate non-glaucomatous trabecular meshwork cellular behavior. Acta biomaterialia 71, 444–459, https://doi.org/10.1016/j.actbio.2018.02.037 (2018).
Dhamodaran, K., Baidouri, H., Sandoval, L. & Raghunathan, V. Wnt activation after inhibition restores trabecular meshwork cells toward a normal phenotype. Investig. ophthalmology I& visual science 61, 30, https://doi.org/10.1167/iovs.61.6.30 (2020).
Yemanyi, F., Vranka, J. & Raghunathan, V. K. Glucocorticoid-induced cell-derived matrix modulates transforming growth factor β2 signaling in human trabecular meshwork cells. Sci. reports 10, 15641, https://doi.org/10.1038/s41598-020-72779-w (2020).
Morgan, J. T., Raghunathan, V. K., Chang, Y.-R., Murphy, C. J. & Russell, P. The intrinsic stiffness of human trabecular meshwork cells increases with senescence. Oncotarget 6, 15362–15374, https://doi.org/10.18632/oncotarget.3798 (2015).
Li, J. et al. Integrated multi-omics single cell atlas of the human retina. https://doi.org/10.1101/2023.11.07.566105 (2023).
Li, J. et al. Comprehensive single-cell atlas of the mouse retina. https://doi.org/10.1101/2024.01.24.577060 (2024).
Heiser, C. N., Wang, V. M., Chen, B., Hughey, J. J. & Lau, K. S. Automated quality control and cell identification of droplet-based single-cell data using dropkick. Genome Res. 31, 1742–1752, https://doi.org/10.1101/gr.271908.120 (2021).
Young, M. D. & Behjati, S. Soupx removes ambient rna contamination from droplet-based single-cell rna sequencing data. GigaScience 9 https://doi.org/10.1093/gigascience/giaa151 (2020).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. Doubletfinder: Doublet detection in single-cell rna sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4, https://doi.org/10.1016/j.cels.2019.03.003 (2019).
Gayoso, A. et al. A python library for probabilistic analysis of single-cell omics data. Nat. Biotechnol. 40, 163–166, https://doi.org/10.1038/s41587-021-01206-w (2022).
Wolf, F. A., Angerer, P. & Theis, F. J. Scanpy: large-scale single-cell gene expression data analysis. Genome Biol. 19 https://doi.org/10.1186/s13059-017-1382-0 (2018).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell https://doi.org/10.1016/j.cell.2021.04.048 (2021).
Miao, Z. et al. Putative cell type discovery from single-cell gene expression data. Nat. Methods 17, 621–628, https://doi.org/10.1038/s41592-020-0825-9 (2020).
Alquicira-Hernandez, J., Sathe, A., Ji, H. P., Nguyen, Q. & Powell, J. E. scpred: accurate supervised method for cell-type classification from single-cell rna-seq data. Genome Biol. 20 https://doi.org/10.1186/s13059-019-1862-5 (2019).
Bunis, D. G., Andrews, J., Fragiadakis, G. K., Burt, T. D. & Sirota, M. dittoseq: universal user-friendly single-cell and bulk rna sequencing visualization toolkit. Bioinformatics 36, 5535–5536, https://doi.org/10.1093/bioinformatics/btaa1011 (2020).
NCBI GEO. https://identifiers.org/geo/GSE263230 (2024).
Schlunck, G. et al. Substrate rigidity modulates cell–matrix interactions and protein expression in human trabecular meshwork cells. Investig. Opthalmology I& Vis. Sci. 49, 262, https://doi.org/10.1167/iovs.07-0956 (2008).
Bagué, T. et al. Effects of netarsudil-family rho kinase inhibitors on human trabecular meshwork cell contractility and actin remodeling using a bioengineered ecm hydrogel. Front. Ophthalmol. 2, https://doi.org/10.3389/fopht.2022.948397 (2022).
Li, H., Raghunathan, V., Stamer, W. D., Ganapathy, P. S. & Herberg, S. Extracellular matrix stiffness and tgf2 regulate yap/taz activity in human trabecular meshwork cells. Front. Cell Dev. Biol. 10, https://doi.org/10.3389/fcell.2022.844342 (2022).
Yoo, H. et al. Simvastatin attenuates glucocorticoid-induced human trabecular meshwork cell dysfunction via yap/taz inactivation. Curr. Eye Res. 48, 736–749, https://doi.org/10.1080/02713683.2023.2206067 (2023).
Li, H. et al. Targeting yap mechanosignaling to ameliorate stiffness-induced schlemm’s canal cell pathobiology. https://doi.org/10.1101/2023.09.08.556840 (2023).
Mitton, K. et al. Transient loss of b-crystallin: An early cellular response to mechanical stretch. Biochem. Biophys. Res. Commun. 235, 69–73, https://doi.org/10.1006/bbrc.1997.6737 (1997).
Tumminia, S. J. et al. Mechanical stretch alters the actin cytoskeletal network and signal transduction in human trabecular meshwork cells. Investig. ophthalmology I& visual science 39, 1361–1371 (1998).
Okada, Y. Bovine trabecular cells produce timp-1 and mmp-2 in response to mechanical stretching. Jpn. J. Ophthalmol. 42, 90–94, https://doi.org/10.1016/s0021-5155(97)00129-9 (1998).
WuDunn, D. The effect of mechanical strain on matrix metalloproteinase production by bovine trabecular meshwork cells. Curr. Eye Res. 22, 394–397, https://doi.org/10.1076/ceyr.22.5.394.5500 (2001).
Bradley, J. M. et al. Effects of mechanical stretching on trabecular matrix metalloproteinases. Investig. ophthalmology I& visual science 42, 1505–1513 (2001).
Bradley, J. M. B., Kelley, M. J., Rose, A. & Acott, T. S. Signaling pathways used in trabecular matrix metalloproteinase response to mechanical stretch. Investig. Opthalmology I& Vis. Sci. 44, 5174, https://doi.org/10.1167/iovs.03-0213 (2003).
Vittal, V., Rose, A., Gregory, K. E., Kelley, M. J. & Acott, T. S. Changes in gene expression by trabecular meshwork cells in response to mechanical stretching. Investig. Opthalmology I& Vis. Sci. 46, 2857, https://doi.org/10.1167/iovs.05-0075 (2005).
Liton, P. B. et al. Induction of il-6 expression by mechanical stress in the trabecular meshwork. Biochem. Biophys. Res. Commun. 337, 1229–1236, https://doi.org/10.1016/j.bbrc.2005.09.182 (2005).
Porter, K. M., Jeyabalan, N. & Liton, P. B. Mtor-independent induction of autophagy in trabecular meshwork cells subjected to biaxial stretch. Biochimica et Biophys. Acta (BBA) - Mol. Cell Res. 1843, 1054–1062, https://doi.org/10.1016/j.bbamcr.2014.02.010 (2014).
Lakk, M. & Križaj, D. Mechanically induced cytoskeletal remodeling in trabecular meshwork cells requires trpv4 - rho signaling interactions. https://doi.org/10.1101/2020.08.11.247171 (2020).
Yarishkin, O. et al. Mechanotransduction and dynamic outflow regulation in trabecular meshwork requires piezo1 channels. https://doi.org/10.1101/2020.06.30.180653 (2020).
Shim, M. S., Nettesheim, A., Dixon, A. & Liton, P. B. Primary cilia and the reciprocal activation of akt and smad2/3 regulate stretch-induced autophagy in trabecular meshwork cells. Proc. Natl. Acad. Sci. 118 https://doi.org/10.1073/pnas.2021942118 (2021).
Russell, P., Gasiorowski, J. Z., Nealy, P. F. & Murphy, C. J. Response of human trabecular meshwork cells to topographic cues on the nanoscale level. Investig. Opthalmology I& Vis. Sci. 49, 629, https://doi.org/10.1167/iovs.07-1192 (2008).
Gasiorowski, J. Z. & Russell, P. Biological properties of trabecular meshwork cells. Exp. Eye Res. 88, 671–675, https://doi.org/10.1016/j.exer.2008.08.006 (2009).
Thomasy, S. M., Wood, J. A., Kass, P. H., Murphy, C. J. & Russell, P. Substratum stiffness and latrunculin b regulate matrix gene and protein expression in human trabecular meshwork cells. Investig. Opthalmology I& Vis. Sci. 53, 952, https://doi.org/10.1167/iovs.11-8526 (2012).
Peng, T. et al. A mathematical model of mechanotransduction reveals how mechanical memory regulates mesenchymal stem cell fate decisions. BMC Syst. Biol. 11, https://doi.org/10.1186/s12918-017-0429-x (2017).
Nasrollahi, S. et al. Past matrix stiffness primes epithelial cells and regulates their future collective migration through a mechanical memory. Biomaterials 146, 146–155, https://doi.org/10.1016/j.biomaterials.2017.09.012 (2017).
Li, C. X. et al. Microrna-21 preserves the fibrotic mechanical memory of mesenchymal stem cells. Nat. Mater. 16, 379–389, https://doi.org/10.1038/nmat4780 (2016).
Jain, S. et al. The role of single-cell mechanical behaviour and polarity in driving collective cell migration. Nat. Phys. 16, 802–809, https://doi.org/10.1038/s41567-020-0875-z (2020).
Mathur, J., Shenoy, V. B. & Pathak, A. Mechanical memory in cells emerges from mechanotransduction with transcriptional feedback and epigenetic plasticity. https://doi.org/10.1101/2020.03.20.000802 (2020).
Lele, T. P., Brock, A. & Peyton, S. R. Emerging concepts and tools in cell mechanomemory. Annals Biomed. Eng. 48, 2103–2112, https://doi.org/10.1007/s10439-019-02412-z (2019).
Scott, A. K., Rafuse, M. & Neu, C. P. Mechanically induced alterations in chromatin architecture guide the balance between cell plasticity and mechanical memory. Front. Cell Dev. Biol. 11, https://doi.org/10.3389/fcell.2023.1084759 (2023).
Espinosa-Martínez, M., Alcázar-Fabra, M. & Landeira, D. The molecular basis of cell memory in mammals: The epigenetic cycle. Sci. Adv. 10, https://doi.org/10.1126/sciadv.adl3188 (2024).
Henikoff, S. & Greally, J. M. Epigenetics, cellular memory and gene regulation. Curr. Biol. 26, R644–R648, https://doi.org/10.1016/j.cub.2016.06.011 (2016).
Killaars, A. R. et al. Extended exposure to stiff microenvironments leads to persistent chromatin remodeling in human mesenchymal stem cells. Adv. Sci. 6, https://doi.org/10.1002/advs.201801483 (2018).
d’Alessandro, J. et al. Cell migration guided by long-lived spatial memory. Nat. Commun. 12, https://doi.org/10.1038/s41467-021-24249-8 (2021).
Balsalobre, A. & Drouin, J. Pioneer factors as master regulators of the epigenome and cell fate. Nat. Rev. Mol. Cell Biol. 23, 449–464, https://doi.org/10.1038/s41580-022-00464-z (2022).
Paro, R., Grossniklaus, U., Santoro, R. & Wutz, A. Cellular Memory, chap. Chapter 3, Cellular Memory, 49–66 (Springer International Publishing, 2021).
Kalukula, Y., Luciano, M., Charras, G., Brückner, D. B. & Gabriele, S. The actin cortex acts as a mechanical memory of morphology in confined migrating cells. https://doi.org/10.1101/2024.08.05.606589 (2024).
Phillips, N. E., Mandic, A., Omidi, S., Naef, F. & Suter, D. M. Memory and relatedness of transcriptional activity in mammalian cell lineages. Nat. Commun. 10, https://doi.org/10.1038/s41467-019-09189-8 (2019).
Elsafi Mabrouk, M. H., Zeevaert, K., Henneke, A.-C., Maaßen, C. & Wagner, W. Substrate elasticity does not impact dna methylation changes during differentiation of pluripotent stem cells. Cytotherapy 26, 1046–1051, https://doi.org/10.1016/j.jcyt.2024.03.485 (2024).
Zhao, X. et al. Extracellular matrix stiffness regulates dna methylation by pkc-dependent nuclear transport of dnmt3l. Adv. Healthc. Mater. 10, https://doi.org/10.1002/adhm.202100821 (2021).
Sumey, J. L., Johnston, P. C., Harrell, A. M. & Caliari, S. R. Hydrogel mechanics regulate fibroblast dna methylation and chromatin condensation. Biomater. Sci. 11, 2886–2897, https://doi.org/10.1039/d2bm02058k (2023).
Lutolf, M. P. et al. In vitro human cell-based models: What can they do and what are their limitations? Cell 187, 4439–4443, https://doi.org/10.1016/j.cell.2024.07.042 (2024).
Morris, S. A. The evolving concept of cell identity in the single cell era. Development 146, https://doi.org/10.1242/dev.169748 (2019).
Gulati, G. S., D’Silva, J. P., Liu, Y., Wang, L. & Newman, A. M. Profiling cell identity and tissue architecture with single-cell and spatial transcriptomics. Nat. Rev. Mol. Cell Biol. https://doi.org/10.1038/s41580-024-00768-2 (2024).
Zeng, H. What is a cell type and how to define it? Cell 185, 2739–2755, https://doi.org/10.1016/j.cell.2022.06.031 (2022).
Savulescu, A. F., Jacobs, C., Negishi, Y., Davignon, L. & Mhlanga, M. M. Pinpointing cell identity in time and space. Front. Mol. Biosci. 7, https://doi.org/10.3389/fmolb.2020.00209 (2020).
Acknowledgements
This project was funded by NIH/NEI R01EY022356 (R.C.), R01EY018571 (R.C.), S10OD032189 (R.C.), R01EY026048-01A1 (V.R.), Chan Zuckerberg Initiative (CZI) award CZF2019-002425, RRF to R.C. It was supported by the Single Cell Genomics Core at Baylor College of Medicine with funding from the CPRIT RP200504. The authors acknowledge support to the Gavin Herbert Eye Institute at the University of California, Irvine from an unrestricted grant from Research to Prevent Blindness and from NIH core grant P30 EY034070. We would also like to thank the Lions Eye Bank (TX), Lions Vision-Gift (OR), and SavingSight (MO) for procuring all human donor eyes used in this work. Most importantly, we would like to thank the families of the organ donors without whose consent these experiments would be impossible. Janice A Vranka passed away in 2021 after a battle with cancer; her intellectual contributions to the study are invaluable.
Author information
Authors and Affiliations
Contributions
A.T., V.R. and R.C conceptualized and designed the study. X.C. and Y.L. generated the scRNA-seq datasets and A.T., S.K. and L.J. preformed data analysis in this study. All authors wrote, reviewed, and contributed to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tian, A., Kim, S., Baidouri, H. et al. Divergence in cellular markers observed in single-cell transcriptomics datasets between cultured primary trabecular meshwork cells and tissues. Sci Data 12, 264 (2025). https://doi.org/10.1038/s41597-025-04528-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04528-5
This article is cited by
-
iPSC-derived ITGA6-positive cells restore aqueous humor outflow in glaucoma eyes
Nature Communications (2025)