Abstract
A remarkable life cycle of the protandrous blackhead seabream (Acanthopagrus schlegelii), initiating as a male during the first two years and then naturally transforming to a female since the third year, makes this fish a valuable model for studying molecular mechanisms of sex change. Here, we constructed a gap-free telomere-to-telomere (T2T) genome assembly for a male blackhead seabream, by integration of PacBio HiFi, Ultra-long ONT and Hi-C sequencing techniques. With 97.87% of the entire sequences anchored into 24 chromosomes, this haplotypic genome assembly spans 714.98 Mb. In terms of correctness (quality value QV: 52.95) and completeness (BUSCO score: 99.9%), this chromosome-scale assembly is indeed of high quality. It has been annotated with 24,581 protein-coding genes, and predicted with low percentage (30.95%) of repetitive sequences. As the first reference T2T-level genome assembly of various protandrous fishes, it provides a valuable genetic resource for expansion of fish genomics database. It will also allow for in-depth genomic comparisons among diverse hermaphrodite vertebrates, as well as offer fundamental genome data to support extensive research on blackhead seabream.
Similar content being viewed by others
Background & Summary
Belonging to the order Perciformes and the family Sparidae, blackhead seabream (Acanthopagrus schlegelii) has been among the most economically important and well-liked marine fish species. It has quick development, outstanding flesh quality, and good environmental adaptability1,2. Its artificial breeding and aquaculture is expanding quickly as a result of the growing demand in the market. Implementing artificial breeding or selection has been employed to facilitate the increased production of blackhead seabream. Sex control is therefore applied as one of the most crucial strategies.
Interestingly, blackhead seabream is a protodromous hermaphrodite with an impressive life cycle of natural sex change from male to female, making it a valuable model for studying molecular mechanisms of fish sex development3.The fish exhibit sexual differentiation during its juvenile stage, possessing a bisexual gonad. This can lead to a fact that male and female reproductions are not synchronized and the ratio of female to male is severely disproportional. Without the ability to regulate sexual differentiation, maturation, and reproduction, farmers indeed have little control over practical breeding processes. Interestingly, previous studies showed that age and season play an combined impact on the dynamic process of bisexual gonad development3. Meanwhile, development of vitellogenic oocytes in the ovary serves as one of the indicators for natural sex change in blackhead seabream4,5. Although some genes such as wnt4 (Wnt family member 4), foxl2 (Forkhead Box L2), cyp19a1a (cytochrome P450, family 19, subfamily A, polypeptide 1a), dmrt1 (doublesex and mab-3 related transcription factor 1), amh (anti-Mullerian hormone), and amhr2 (anti-Mullerian hormone receptor type 2), potentially related to sexual differentiation and sex controlling, have been reported1,4,5; however, the detailed molecular mechanisms of natural sex change is still unknown in blackhead seabream.
For investigations of functional, ecological, and evolutionary genomics in this species as well as other hermaphrodite fishes, it is necessary for researchers to have a well-assembled genome in hand. A draft genome assembly of a female blackhead seabream was released by us in 2018, containing 89% of the full BUSCOs (actinopterygii_odb9 database) and numerous contigs (115, 091) with a low N50 value of 17.2 kb6. Although this assembly provided an important genetic resource for comparative genomics studies on various Perciformes, its fragmentation and incompleteness has restricted its comprehensive applications in fish research. In our present work, we assembled a gap-free telomere-to-telomere (T2T) genome of a male blackhead seabream by integration of PacBio (Pacific Biosciences) HiFi long reads, ONT (Oxford Nanopore Technologies) ultra-long reads, MGI short reads, and Hi-C (High-through chromosome conformation capture) sequencing reads. This high-quality genome dataset will greatly enable further works on understanding of biological characteristics (especially sex change) of blackhead seabream.
Methods
Sample collection
A two-year-old male adult blackhead seabream (Fig. 1a) was collected from Guangdong Marine Fisheries Experimental Centre, which belongs to the Agro-Tech Extension Center of Guangdong Province with an offsite location in Huizhou city, Guangdong province, China. Muscle tissue was pooled for whole genome sequencing, and muscle, liver, brain, gill, and gonad were collected for additional transcriptome sequencing. The sampling procedure and practical pipeline were conducted according to the recommendations and approval of the Animal Ethics Committee of Shenzhen University (Shenzhen, China; license number: A202301455).
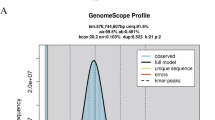
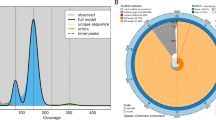
A T2T genome assembly of the protandrous hermaphrodite blackhead seabream. (a) A photo of the sequenced fish. (b) A GenomeScope k-mer plot. (c) A total of 24 distinct blocks in the Hi-C contact matrixes. (d) A Circos plot of the main genome features. From outside to inside: (I) the 24 chromosomes, (II) repeats, (III) Long terminal repeat (ltr), (IV) gene density, and (V) GC content. Links inside the Circos refer to internal syntenic blocks among different chromosomes within the assembled genome.
DNA extraction and genome sequencing
A blood & cell culture DNA kit (Qiagen, USA) was used for extraction and purification of genomic DNA (gDNA) from the muscle in accordance with the manufacturer’s instructions. The extracted gDNA was used for construction of a library (insert size of 350 bp) with the MGIEasy Universal DNA Library Prep Kit (MGI, China), which was then sequenced on an MGISEQ. 2000 platform (MGI). A total of 100.47 Gb of raw reads (150 bp in length) were generated, among them low-quality reads and adaptor sequences were filtered using SOAPfilter (v2.2)7 with default settings. Finally, we obtained 95.98 Gb of clean reads for estimating genome size and assembling sequences.
Additionally, long-read libraries were created utilizing a PacBio Sequel II System and a SMRTbell Express Template Prep Kit 2.0 for HiFi sequencing, following PacBio’s standard technique (Pacific Biosciences, USA). The consensus sequences were then produced using the CCS software (SMRT Link v9.0)8. Approximately 62.60 Gb of consensus reads with a mean length of 15.25 kb were obtained.
Oxford Nanopore Technologies (ONT) was applied for construction of an ultra-long library and then one flow cell was sequenced on a PromethION platform (Oxford Nanopore Technologies Co., UK). The raw reads were first refined to remove those with quality value (QV) below 7. Subsequently, Porechop (https://github.com/rrwick/Porechop) was applied to eliminate adaptors, and Filtlong (https://github.com/rrwick/Filtlong) was employed to filter out those reads shorter than 30 kb and mean read quality scores less than 90%. Finally, a total of 1.171 million clean reads were retained, accumulating a substantial count of 22.44 Gb. The average read length was 79.63 kb, with an N50 length of 100 kb.
For the Hi-C sequencing, DNA libraries were prepared with a GrandOmics Hi-C kit (GrandOmics, China; using DpnII as the restriction enzyme) as per the manufacturer’s instructions. The Hi-C libraries were then sequenced on an Illumina Novaseq system (Illumina, USA), generating 76.46 Gb of 150-bp paired-end raw reads. Trimmomatic (V0.39)9 with optimized parameters “SLIDINGWINDOW: 4:20, LEADING: 3, TRAILING:3, ILLUMINACLIP: adapter.fa:2:30:10:8:true” was used to filter out adaptor sequences, low-quality reads (quality scores < 20), and those reads shorter than 36 bp. This filtering process retained 63.53 Gb of clean data for subsequent construction of chromosomes.
RNA extraction and transcriptome sequencing
Using a normal Trizol methodology (Invitrogen, USA), RNA samples were isolated from muscle, liver, brain, gill, and gonad tissues, and then purified using a Qiagen RNeasy Mini Kit (Qiagen). Following the manufacturer’s instructions, equivalent amount of RNA from each tissue were combined to create an Illumina cDNA library, which was subsequently sequenced on a HiSeq X Ten platform (Illumina). After generating 31.12 Gb of paired-end raw reads, adaptor sequences and low-quality reads were removed using SOAPfilter (v2.2)7 with default settings. In the end, 27.1 Gb of clean reads were retained for annotation of gene structures.
Genome assembly
Genome-size estimation
Using Jellyfish (v2.2.6)10 and GenomeScope (v2.0)11, a K-mer frequency distribution of the MGI clean reads was determined. Accordingly, the genome size of blackhead seabream is calculated to be 671.32 Mb, and the rate of genomic heterozygosity was predicted to be 0.59% (Fig. 1b).
De novo genome assembly
In this study, HiFiasm (v0.19.5)12 was employed for assembling into contigs using HiFi + ONT long reads. Subsequently, these contigs were refined by T2T-polish13 with the optimized parameter set to task = best using the MGI short reads.This primary genome assembly was 731.09 Mb in length, and it is anchored into 41 contigs (with a contig N50 of 31.8 Mb).
Construction of chromosomes and gap filling
This high-quality genome assembly served as the foundation for subsequent construction of chromosomes using the Hi-C reads. Initially, Bowtie2 (v2.3.2)14 was employed to map clean reads to the primary genome assembly. Subsequently, the HiC-Pro (v2.8.1)15 pipeline was employed to detect valid contact paired reads. Using these Hi-C valid reads, the assembled contigs were anchored onto chromosomes via the 3D-DNA pipeline16 with the parameter set to -r 0. Manual correction of the chromosome-level scaffolds was then performed using JuiceBox (v1.11.08)17. Based on the HiFi and ONT long reads, we applied TGS-GapCloser (v1.1.1)18 with default parameters to resolve any remaining gaps in the chromosome-level genome assembly. As a result, we obtained the final genome assembly, which has a total size of 714.71 Mb and is anchored into 24 chromosomes with 97.78% of the primary assembly sequences (Fig. 1c, d and Table 1). Its contig N50 reaches 31.8 Mb.
Identification of centromere and telomere sequences
We identified telomere sequences through searching for repeating sequences (TTAGGG/CCCTAA) in telomeric regions. Centromere identification was performed by using the Centromics program (https://github.com/ShuaiNIEgithub/Centromics), which dealt with raw HiFi sequencing data, Hi-C sequencing data, and genome assembly data. Ultimately, we discovered that blackhead seabream chromosomes owned 24 centromeres and 47 telomeres (see more details in Fig. 2 and Table 1).
Genome annotation
Repetitive sequence annotation
Both ab initio and homology-based strategies were employed to detect repetitive sequences in the blackhead seabream genome. In detail, using RepeatModeler (v2.0.1)19 and MITE-Hunter20 program with default settings, we constucted an ab initio repeat sequence library. This library was then aligned to the Repbase 24.021 for classification of different repetitive elements via the TEclass tool22. For the homolog-based prediction based on the Repbase database21, DNA and protein transposable elements (TEs) were detected by RepeatMasker (v4.1.2) and RepeatProteinMasker (v4.0.7)23, respectively. Tandem repeats were identified with Tandem Repeat Finder (v4.10.0)24. After removal of redundant data from both methods, our results showed that a total of 221.23 Mb of repetitive sequences were identified, accounting for 30.95% of the assembled blackhead seabream genome (Table 2).
Protein-coding genes and functional annotations
Three methods were combined to annotate protein-coding genes, including ab initio gene prediction, homology-based annotation, and transcriptome-based annotation. Using the HISAT2 (v2.2.1)25, refined transcriptome (RNA-seq) reads were aligned to the assembled genome for the transcriptome-based annotation. PASA26 was then applied to detect open reading frames (ORFs), and Stringtie27 was used for gene structure annotation by assembling corresponding transcripts. For the homology prediction, GeMoMa (v2.3)28 was employed to map protein sequences from four representative species (including yellowfin seabream Acanthopagrus latus, sharksucker Echeneis naucrates, zebrafish Danio rerio, and large yellow croaker Larimichthys crocea) to our assembly for prediction of gene structures. For the ab initio prediction, a training model library was constructed from a protein set generated from the RNA-seq reads by Trinity (v2.8.5)29. Augustus (v3.4.0)30 was then employed to annotate genes with the–noInFrameStop = true–strand = both setting based on the training data.
Integration of protein-coding genes annotated by these three approaches was carried out using the EVidenceModeler (EVM) pipeline (v1.1.1)26. Consequently, 24,081 protein-coding genes were identified, with an average gene length of 16.34 kb and an average coding sequence (CDS) length of 1,721.43 bp. The average number of exons per gene was 10.28, while exons averaged 271.91 bp in length and introns averaged 1,437.93 bp (see Table 3).
The protein-coding genes were functionally annotated by aligning them with several routine protein databases using Blastp (v2.2.26)31 and Diamond (v2.0.7)32. This alignment included comparisons with the NCBI nonredundant (NR) protein database (v5; released on September 29, 2020), as well as Swissprot (released on October 07, 2020)33, KEGG (released on October 1, 2019)34, TrEMBL (released on October 07, 2020)34, InterPro (v5.50–84.0)35 and Gene Ontology (GO; v1.2, released on July 27, 2023)36 databases. Of the total gene models, 24,179 (98.36%) were annotated with at least one homologous hit from these public databases (refer to Table 3).
Annotation of non-coding RNA genes
For annotation of non-coding RNA genes, tRNAscan-SE (v2.0.9)37 with default settings was employed to annotate tRNA-associated genes; rRNAs annotation was carried out using RNAmmer (v1.2)38; miRNAs and snRNAs were detected using Infernal (v1.1.2)39 against the Rfam (v1.4.1) database40 with default parameters. As a result, annotations predicted 956 rRNAs, 2,175 tRNAs, 874 miRNAs, and 1,212 snRNAs (see more details in Table 4).
Data Records
All genome data have been uploaded to the NCBI SRA database with the BioProject accession PRJNA1134337, including the specific accessions SRR29908548 to SRR2990855141,42,43,44. The genome assembly have been deposited in the GenBank database under the accession number JBGQWW00000000045. Furthermore, detailed documents on genome assembly, gene structures, gene functions, and repeat annotations for blackhead seabream have been shared on the Figshare46.
Technical Validation
Evaluation of the genome assembly and annotation
To evaluate the quality of our genome assembly, we employed several approaches. First, we employed BUSCO (v5.2.2)47 to examine completeness. The BUSCO analysis revealed that, based on the 3640 single-copy orthologs in the actinopterygii_odb10 database, 99.3% of our annotated genes were correctly classified as complete (99.1% single-copy genes and 0.2% duplicated genes), with 0.6% being fragmented (see Table 5). Second, Merqury (v1.328)48 estimated the assembly quality value (QV) to be 52.95. Third, by aligning the sequencing data to the assembled genome, we evaluated the accuracy rate, which showed mapping rates of 96.53% for RNA-Seq data, 99.43% for the MGI data, 99.74% for the PacBio data, and 99.99% for the ONT data. These results collectively indicate high quality of the blackhead seabream genome assembly. Moreover, a BUSCO analysis was performed to assess the completeness of the gene structure annotation, revealing that 96.2% of our annotated genes were correctly classified as complete, with 0.8% being fragmented.
Collinearity analysis
Whole-genome synteny analysis was performed using the GenomeSyn (v1.2.7)49 by aligning the chromosome-level genome assemblies between blackhead seabream (this study) and its relative yellowfin seabream (A. latus)50. Our results prove that they had excellent one-to-one correspondences among their chromosomes (Fig. 3). This good similarity also underlines the high quality of our sequencing and assembly of the blackhead seabream genome.
Good synteny of chromosomes between blackhead seabream (Acanthopagrus schlegelii) and its relative yellowfin seabream (Acanthopagrus latus)46.
Code availability
No custom code was used for this study. The versions and parameters for employed software have been deposited in Figshare (https://doi.org/10.6084/m9.figshare.26362411.v5). Whenever particular parameters were missing for a software type, the default settings suggested by the creators took effect.
References
Lin, Z. et al. Comparative transcriptome analysis of mixed tissues of black porgy (Acanthopagrus schlegelii) with differing growth rates. Aquac. Res. 52, 5800–5813 (2021).
Zhang, K. et al. A comparative transcriptomic study on developmental gonads provides novel insights into sex change in the protandrous black porgy (Acanthopagrus schlegelii). Genomics 111, 277–283 (2019).
Lee, M.-F., Huang, J.-D. & Chang, C.-F. Development of ovarian tissue and female germ cells in the protandrous black porgy, Acanthopagrus schlegeli (Perciformes, Sparidae). Zool. Stud. 47, 302–316 (2008).
Wu, G. C. et al. Sex differentiation and sex change in the protandrous blackhead seabream, Acanthopagrus schlegeli. Gen. Comp. Endocrinol. 167, 417–421 (2010).
Wu, G., Dufour, S. & Chang, C. Molecular and Cellular Endocrinology Molecular and cellular regulation on sex change in hermaphroditic fish, with a special focus on protandrous black porgy, Acanthopagrus schlegelii. Mol. Cell. Endocrinol. 520, 111069 (2021).
Zhang, Z. et al. Draft genome of the protandrous Chinese black porgy, Acanthopagrus schlegelii. Gigascience 7, giy012 (2018).
Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 2047–217X (2012).
Rhoads, A. & Au, K. F. PacBio sequencing and its applications. Genom. Proteom. Bioinfor. 13, 278–289 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Ranallo-Benavidez, T. R., Jaron, K. S. & Schatz, M. C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432 (2020).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with HiFiasm. Nat. Methods 18, 170–175 (2021).
Cartney, A. M. et al. Chasing perfection: validation and polishing strategies for telomere-to-telomere genome assemblies. Nat Methods 19, 687–695 (2022).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 1–11 (2015).
Durand, N. C. et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 3, 95–98 (2016).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016).
Xu, M. et al. TGS-GapCloser: fast and accurately passing through the Bermuda in large genome using error-prone third-generation long reads. BioRxiv 831248 (2019).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinforma. 1–14 (2009).
Xu, Z. & Wang, H. LTR-FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, 265–268 (2007).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. Dna 6, 1–6 (2015).
Abrusán, G., Grundmann, N., DeMester, L. & Makalowski, W. TEclass—a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25, 1329–1330 (2009).
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21, i351–i358 (2005).
Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360 (2015).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9, 1–22 (2008).
Kovaka, S. et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 1–13 (2019).
Keilwagen, J. et al. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 44, e89–e89 (2016).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Stanke, M. & Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33, 465–467 (2005).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Bairoch, A. & Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48 (2000).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Finn, R. D. et al. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199 (2017).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Lagesen, K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108 (2007).
Nawrocki, E. P., Kolbe, D. L. & Eddy, S. R. Infernal 1.0: inference of RNA alignments. Bioinformatics 25, 1335–1337 (2009).
Kalvari, I. et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 49, D192–D200 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR29908548 (2024).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR29908549 (2024).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR29908550 (2024).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR29908551 (2024).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_041753875.1 (2024).
Zhang, K. Acanthopagrus schlegelii annotation files. figshare https://doi.org/10.6084/m9.figshare.26362411.v5 (2024).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21, 1–27 (2020).
Zhou, Z. et al. GenomeSyn: a bioinformatics tool for visualizing genome synteny and structural variations. Journal of genetics and genomics 49, 1174–1176 (2022).
Lu, J. et al. Chromosome-level genome assembly of Acanthopagrus latus provides insights into salinity stress adaptation of Sparidae. Mar. Biotechnol. 24, 655–660 (2022).
Acknowledgements
This project was supported by National Key R & D Program of China (No. 2020YFA0908700 and 2022YFE0139700), Guangdong Basic and Applied Basic Research Foundation (No. 2023A1515110554), and Research Initiation Fund for Young Faculty Members at Shenzhen University (No. 000001032214).
Author information
Authors and Affiliations
Contributions
L.D. and Q.S. conceived this study. K.Z. performed data analysis; J.W., S.Y. and S.G. participated in the collection of samples; W.Z. and X.Z. provided research advice; K.Z. wrote the draft manuscript. Q.S. and L.D. revised the manuscript. All authors have read and approved the final manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, K., Guo, S., Yang, S. et al. A telomere-to-telomere genome assembly of the protandrous hermaphrodite blackhead seabream, Acanthopagrus schlegelii. Sci Data 12, 350 (2025). https://doi.org/10.1038/s41597-025-04602-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04602-y