Abstract
Hyperspectral imaging (HSI) and machine learning (ML) have been employed in the medical field for classifying highly infiltrative brain tumours. Although existing HSI databases of in vivo human brains are available, they present two main deficiencies. First, the amount of labelled data are scarce, and second, 3D-tissue information is unavailable. To address both issues, we present the SLIMBRAIN database, a multimodal image database of in vivo human brains that provides HS brain tissue data within the 400–1000 nm spectra, as well as RGB, depth and multiview images. Two HS cameras, two depth cameras and different RGB sensors were used to capture images and videos from 193 patients. All the data in the SLIMBRAIN database can be used in a variety of ways, for example, to train ML models with more than 1 million HS pixels available and labelled by neurosurgeons, to reconstruct 3D scenes or to visualize RGB brain images with different pathologies, offering unprecedented flexibility for both the medical and engineering communities.
Similar content being viewed by others
Background & Summary
Neuroscience databases commonly provide information regarding gene expression1,2,3, neurons4,5, macroscopic brain structure6,7,8,9, and neurological or psychiatric conditions10,11,12,13, which provides researchers with a better understanding of human and animal brains. Most imaging techniques acquire information without the need to make an incision in the subject. Although this approach can provide useful biological information, there is a need to provide intraoperative tools for neurosurgeons during surgical procedures. In fact, delineating pathological tissue from healthy tissue is a difficult task for neurosurgeons when removing brain tumours. Moreover, glioblastoma multiforme (GBM) is one of the most common and aggressive tumours, as long-term survival is not guaranteed14, and resection is extremely difficult owing to the high infiltration capabilities of GBM15. In most cases, surgical intervention is an unavoidable step to increase patient survival. Furthermore, once the brain is exposed after craniotomy, it derepressurizes and cerebrospinal fluid leaks, causing the brain to shift towards the surgical opening16. Existing tools help neurosurgeons remove as many tumours as possible while preserving healthy intact tissue. These tools include neuronavigators, intraoperative magnetic resonance (iMR), intraoperative ultrasound (IOUS) or add-on agents such as 5-aminolevunilic acid (5-ALA). Although they are commonly used, we now describe their advantages and weaknesses. First, neuronavigation tools17 require magnetic resonance imaging (MRI) prior to surgery to identify the tumour during surgical intervention; however, localization can be difficult once the brain is exposed owing to the brain shift. Second, iMR images solve the previous issue, but MRI-compatible surgical equipment18 is needed, which increases the duration of surgical intervention19. Third, solving the brain shift problem while reducing the time of the operation with surgical tools could be achieved with IOUS20, but these images are resolution-limited and usually present artefacts21. Finally, 5-ALA22 can help solve most issues presented at once, although at the expense of being an invasive method that can be used only for high-grade tumours in adult patients23. Therefore, it is crucial to employ noninvasive techniques that are faster to apply than the tools described above. A widely used technique applied in the medical field is HSI24, which is a noninvasive and nonionizing tool requiring no contact with the patient. Recently, HSI has been used as an intraoperative tool to delineate GBM tumours from in vivo human brains25. Captured images from a previous study have been published in the HELICoiD (HypErspectraL Imaging Cancer Detection) database26, which can provide useful information about several brain tissues in the visible and near infrared (VNIR) spectra. Additionally, it has proven to be useful for tissue classification using ML approaches during surgical interventions27. HELICoiD images are captured with an HS linescan camera, which obtains a single spatial line of pixels with all the spectral information. This requires a scanning procedure that takes at least 1 minute27 and can provide only a single static brain classification image. Thus, real time solutions, understood as processing and classifying a sequence of HS images to provide a live classification video, cannot be achieved with the proposed intraoperative tool. Accordingly, HS snapshot cameras need to be employed to achieve real time classification videos. These kinds of HS cameras obtain the entire spatial and spectral information within a single frame.
In this paper, we present the SLIMBRAIN database, a multimodal image database of in vivo human brains acquired with several HS cameras, RGB cameras and depth sensors. The SLIMBRAIN database contains 193 captures that were obtained during surgical interventions at the Hospital Universitario 12 de Octubre in Madrid, Spain, starting in December 2019. For convenience, the term “capture” is defined as a specific moment in time during the intervention when data acquisition of images is conducted using all sensors. The SLIMBRAIN database has several potential applications. For example, images obtained with different types of sensors can be fused to enhance the classification of brain tissues using HSI and ML. This fusion can be superimposed over generated 3D brain scenes with multiview or depth images28. Additionally, depth, RGB and classification images can be merged in real time to provide live videos of 3D scenes to help neurosurgeons identify the tumours. Another potential use is the characterization of brain tissues from the information provided with any of the two available HS cameras, a linescan covering the VNIR (400–1000 nm) spectra and a snapshot camera measuring fewer spectral bands in the near infrared (NIR) range (660–950 nm)29. Furthermore, labelled hyperspectral data can also be used to develop sophisticated algorithms to enhance brain tumour classification30, compare and evaluate different ML approaches31,32, enhance classification performance in hyperparameter optimization techniques33, segment blood vessels from low-resolution hyperspectral data34, examine the impact of varying the ground truth preprocessing, from sparse to dense35, or combine the data with other databases for the development of robust classification algorithms36. Although SLIMBRAIN can be utilized for engineering purposes, it can also provide helpful knowledge to neuroscience students and researchers. This is because not only preprocessed HS data and depth images but also raw RGB images of in vivo human brains suffering from several pathologies are provided, allowing the visualization of exposed brain surfaces.
Methods
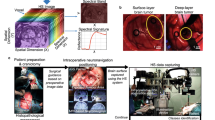
The SLIMBRAIN database provides in vivo human brain images obtained from living patients during surgery. An overview of all methods developed to acquire the SLIMBRAIN database is presented in Fig. 1. At the time of this writing, more than 193 human brains have been captured with several technologies, including HSI, laser imaging detection and ranging (LiDAR) and standard RGB imaging. All these images have been acquired with multiple intraoperative acquisition systems, which have progressively improved over the years. These systems are described in further detail in the “Development of an intraoperative acquisition system” subsection. With all the developed acquisition systems, a repeated procedure was subsequently performed with every patient. This process starts with preparing the patient before surgery and positioning them properly to conduct the surgery, as indicated in Fig. 1 as step 1. Once the brain is exposed, the data acquisition in step 2 is performed with the sensors included in the acquisition system. Later, in step 3, a neuropathological assessment is performed with the resected pathological tissue to confirm the pathology the patient is experiencing. In the meantime, the acquired HS images are processed with a preprocessing chain, which is explained in further detail in step 4. Next, in step 5, these HS images are labelled by neurosurgeons with a labelling tool already used in the state of the art26. The tool allows us to obtain what is considered a ground-truth (GT) map in this study. Finally, with step 6, the labelled data are extracted using the GT and its corresponding preprocessed HS image.
Ethics
The records available in the SLIMBRAIN database were obtained from female and male human patients over 18 years of age who underwent a craniotomy procedure to expose the brain surface. Patients who suffered from any pathological disease or other pathologies other than intrinsic brain tumours (such as incidental aneurysms, incidental arteriovenous malformations, cavernomas, meningiomas or brain metastasis) have been included in the data records. Image collection was carried out from December 2019 onwards at the Hospital Universitario 12 de Octubre in Madrid, Spain. The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Research Ethics Committee of Hospital Universitario 12 de Octubre, Madrid, Spain (protocol code 19/158, 28 May 2019). Informed consent was obtained from all the subjects involved in the study. To anonymously identify each patient, the notation used is based on the “ID” suffix followed by a six-digit number (i.e., ID000023). Both the Patient Information and the Informed Consent documents were approved by the Research Ethics Committee with medicines of Hospital Universitario 12 de Octubre. The Ethics committee approved the procedures for this study on 28 May 2019 with the CEIm code 19/158, and principal investigator was Dr Alfonso Lagares. The patients were explicitly informed in the Patient Information document that images taken outside the usual clinical practice will be part of a database where the patient-related data will be anonymized.
Development of an intraoperative acquisition system
The SLIMBRAIN prototype version 1 was composed of an HS snapshot camera (Ximea GmbH, Münster, NRW, Germany) attached to a tripod. The camera was the first generation of HS snapshot cameras built by Ximea, which captures 25 spectral bands in the 660–950 nm spectral range. Initially, the camera was triggered with the software provided by the manufacturer, but custom software was subsequently developed in Python to obtain HS images. Attached to the camera body, a VIS-NIR fixed focal length lens of 35 mm from Edmund Optics (Edmund Scientific Corporation, Barrington, NJ, USA) was included with a Thorlabs FELH0650 longpass filter (Thorlabs, Inc., Newton, NJ, USA). This filter has a cut-off wavelength of 650 nm. Furthermore, the focus procedure was manually performed by adjusting the focus lens by hand depending on the working distance. Moreover, an external light source with a 150 W halogen lamp (Dolan-Jenner, Boxborough, MA, USA) and a dual-gooseneck fibre optic were integrated into the system to illuminate the brain surface (Dolan-Jenner, Boxborough, MA, USA). This fibre optic illumination covered the spectral range between 400–2200 nm. In addition, a laser rangefinder (TECCPO, Shenzhen, Guangdong, China) meter was used to measure the distance between the HS camera and the brain surface and the angle of the camera with respect to the horizontal. Additional RGB images were captured with mobile phones. These images help neurosurgeons during the labelling process, as they are used as a reference for what they saw during the intervention.
The SLIMBRAIN prototype version 1 was subsequently upgraded to version 2 by replacing the tripod with a scanning platform consisting of an X-LRQ300HL-DE51 motorized linear stage with 300 mm of travel and 0.195 μm precision (Zaber Technologies Inc., Vancouver, BC, Canada). This linear stage was coupled to a horizontal mast attached to a vertical metal support with wheels. In addition to the HS snapshot camera, an MQ022HG-IM-LS150-VISNIR HS linescan camera (Ximea GmbH, Münster, NRW, Germany) was added, which is able to capture 150 effective spectral bands with a 3 nm spacing in the 470–900 nm spectral range with a spatial resolution of 2048 × 5 pixels per band. This camera was configured to capture images with an exposure time of 80 ms. No frame rate was set because the camera was triggered by software to capture a single frame when indicated. A VIS-NIR fixed focal length lens 35 mm from Edmund Optics (Edmund Scientific Corporation, Barrington, NJ, USA) was attached to the camera body. Additionally, the laser used in the SLIMBRAIN prototype version 1 was replaced with the VL53L0X time-of-flight (ToF) distance ranging sensor to automate the process of measuring without needing manual intervention, as with the SLIMBRAIN prototype version 1. The scanning procedure for the linear stage and the data acquisition for both the HS linescan and the snapshot cameras were triggered with software custom developed in C++ programming language. RGB images were also captured with mobile phones.
The SLIMBRAIN prototype was subsequently upgraded to version 3, adding flexibility with height and tilt movements that versions 1 and 2 lacked. The SLIMBRAIN prototype version 3 is presented in Fig. 2(a),(b), which shows the most relevant imaging sensors used and attached to a motorized linear stage. The SLIMBRAIN prototype version 3 included a vertical linear rail with a motor attached (OpenBuilds Part Store, Zephyrhills, FL, USA). The horizontal mast from the SLIMBRAIN prototype version 2 was removed, and a sophisticated system for tilting was used instead. The horizontal motorized linear stage presented in Fig. 2(a),(b) is capable of covering an effective capture range of 230 mm. Although not visible in Fig. 2(a),(b), a vertical rail behind the motorized linear stage can move downwards and upwards in an effective range of 85 cm. A tilting system not visible in the figures but attached to the vertical rail was developed to tilt the horizontal motorized linear stage from 15– to 80 degrees with respect to the camera lenses. The tilt angles are considered 0° and 90° when the camera lenses point to a wall and its floor, respectively. Furthermore, a servomotor with 3D-printed gears was used to adjust the focus lens of the HS snapshot camera, which can be seen in the white case containing the snapshot HS camera and the servomotor gears on the left of Fig. 2(a). To control all three motors that allow the cameras to move horizontally and, vertically and to tilt, as well as the focus procedure and data capture, a C++ custom software with a graphic user interface (GUI) was used. With the described improvements included in the SLIMBRAIN prototype version 3, the system was more flexible than prototypes versions 1 and 2 when images were acquired. Although the same light source and HS snapshot camera were used, the HS linescan camera included in the SLIMBRAIN prototype version 2 was replaced with a Hyperspec® VNIR E-Series linescan camera, which has higher spatial and spectral resolutions capable of capturing 1600 pixels per line and 394 spectral bands in the 365–1005 nm spectral range (Headwall Photonics Inc., Fitchburg, MA, USA). Nonetheless, not all these spectral bands are useful, and only 369 effective spectral bands can be utilized as specified by the manufacturer. Therefore, the useful spectral information gathered with the sensor covers the 400–1000 nm spectral range. Because this camera captures single lines and is based on the pushbroom technique, positioning the camera in the motorized linear stage was suitable for moving it to scan multiple lines and compose brain captures. The motorized linear stage is always configured to move at the appropriate speed to correctly join the all captured lines. The lens used with the linescan camera is a Xenoplan with a maximum aperture of f1.9 (Schneider Kreuznach, Bad Kreuznach, Germany) and a 35 mm focal length. In addition, a Midopt L390 longpass filter (Midwest Optical Systems, Palatine, IL, USA) was placed in front of the lens to block ultraviolet (UV) light and allow > 90% light transmission in the visible and near infrared spectra. Although a servomotor was used for the HS snapshot camera to focus the lens, this was not the case with the HS linescan camera. The reason for this was the difficulty in determining if a single line is in focus in the operating room. Therefore, the lens was focused at a fixed distance of approximately 44 cm. Moreover, a LiDAR device was used to acquire depth maps of the brain scene. It also replaced the necessity of using mobile phones to obtain RGB images, as the LiDAR can obtain RGB images. Specifically, an Intel RealSense L515 (Intel Corporation, Mountain View, CA, USA) was utilized. In addition, the LiDAR allowed the angle and distance at which the captures were taken to be measures. Depth images and videos are monochromatic with 16 bits and 1024 × 768 pixels, whereas RGB images and videos have 8 bits per channel and 1920 × 1080 pixels.
Finally, the newest prototype to date and used to acquire images is the SLIMBRAIN prototype version 4, which, for convenience, will be described in future works in further detail. Nonetheless, the main imaging systems included in the prototype are now described. The LiDAR from the SLIMBRAIN prototype version 3 has been replaced by an Azure Kinect DK (Microsoft Corporation, Redmond, WA, USA) depth camera because Intel L515 production was discontinued. The Azure Kinect camera can obtain the same information as the Intel L515 but with higher spatial resolution in the RGB images. These RGB images are now acquired in ultrahigh definition (UHD) with 3840 × 2160 pixels and 8 bit depth for each of the four channels stored (RGBA). Depth images and videos are acquired with 16 bits and a spatial resolution of 640 × 576 pixels. Although the resolutions of the depth images captured with the Azure Kinect camera are lower than those obtained with the Intel L515 LiDAR, the precision is greater. Additionally, the actual resolution of the depth sensor can be configured to provide 1024 × 1024 pixels, but a lower resolution has been used to keep the frame rate of the camera to 30 frames per second (FPS). Another benefit of the Azure Kinect is that, unlike the Intel L515 LiDAR, which uses its own distance unit and requires multiplication by a constant to retrieve the data in millimetre, the Azure Kinect directly returns the depth measurements in millimetre. Nonetheless, one inconvenience of the Azure Kinect is the reduction in spatial resolution in the depth sensor, as depth images are captured in a rhombus shape. The estimated cost of the SLIMBRAIN prototype version 4, in terms of imaging sensors, including both hyperspectral cameras and the Azure Kinect, is approximately €70,000.
Importantly, different lamps with distinctive spectral responses are used. The spectral responses of the different lamps as well as the ambient fluorescent lighting in the surgery room are shown in Fig. 3 with normalized digital counts for comparison purposes.
Spectral responses of all three lamps (A), (B) and (C) used during the acquisition of images and the ambient fluorescent light in the surgery room (lamp D). The intensity measurements are performed using a spectrometer coupled to a fibre optic pointing directly to the lamps connected to the same power source. The visible (VIS) spectra is illustrated with rainbow shading in the 380–740 nm range, whereas the near ultraviolet (NUV) and near infrared (NIR) spectra are located on the left and right sides, respectively. The spectral ranges of the HS cameras are also presented and illustrated with line arrows above the plot.
A miniature spectrometer capable of measuring in the VIS and near-infrared (NIR) spectra from 350–925 nm (Ocean Insight, Orlando, FL, USA) with an analogue-to-digital resolution of 16 bits was used to analyse the spectral responses of all sources of illumination. The Ocean View program from Ocean Insight was used for this investigation. These findings were attained by using a fibre optic coupled to a spectrometer to measure the light directly emitted from the light lamp. Note that lamps A, B and C from Fig. 3 have been connected to the same external light source previously described. Furthermore, the ambient fluorescent tubes described as lamp D in Fig. 3 are fixed and distributed in the ceiling of the operating room. Lamp A was the first lamp used, which was a 150 W, 21 V EKE lamp included with the external light source by Dolan Jenner, shown in blue in the plot. Then, another two 150 W, 21 V EKE lamps (Osram GmbH, Münich, Freistaat Bayern, Germany) and (International Light Technologies Inc., Peabody, MA, USA), lamps B and C, respectively, replaced lamp A. Lamp B with an orange curve was the first replacement, and then lamp C, with a green response, is shown in Fig. 3. The replacement of lamp A was due to its inconsistency in the 700–925 nm spectra, with very pronounced peaks and valleys. Although lamp B smoothed the response in the previous range, lamp C from International Light (model L1090) replaced both, as it provides more consistent energy within the NIR spectra. In the Data Records section, further information is provided regarding which patients have been illuminated with which lamp. Moreover, the fluorescent background lighting used in the operating room was measured and is also illustrated as lamp D with a red line in Fig. 3. As shown in the figure, very pronounced peaks can be seen in the VIS spectra that have influenced the measurements performed with some hyperspectral cameras. This is important to note, as the lights of the operating room had not been switched off when most of the images in the database are acquired here.
Step 1. Patient preparation
Prior to surgery and in most cases, computed tomography (CT) and MRI of the head of the patient were performed. These images are appropriate for the image-guided stereotactic (IGS) system used by neurosurgeons in the operating room, which helps to determine the location of the tumour in the brain. Both CT and MRI data are uploaded to the IGS system prior to surgery. On the day of the operation, the patient is taken to the operating room and placed in a prone, supine, or lateral recumbent position on a bed under general anaesthesia. Once the patient is ready, their position is registered in the IGS system. Afterwards, hair is removed to allow for the scalp incision to be made, and burr holes are drilled with a high-speed drill. A craniotomy is subsequently performed using a cranial drill, which is inserted into the burr holes to extract a cut bone flap. Finally, a durotomy is performed by cutting the dura with special scissors and exposing the brain surface.
Step 2. Data acquisition
Before entering the operating room, researchers assign patient identifiers based on previous surgeries. Once the brain surface is exposed and ready to be captured, surgical assistants notify the researchers to enter the room with any of the described SLIMBRAIN prototypes. Then, neurosurgeons indicate which zone should be placed in focus and then remove the surgical light to avoid influencing the spectra gathered with the cameras. Immediately, the SLIMBRAIN prototype is positioned next to the patient to proceed with the data acquisition. Importantly, the system must be moved to avoid touching sterile material in the room. Once the system is in place, the acquisition procedure to capture in vivo brain images is illustrated in Fig. 4 and detailed below. Note that this description is provided with the SLIMBRAIN prototype version 3 (Fig. 2(a)) instead of version 4.
-
1.
Turn on the light source to illuminate the brain surface and ask the neurosurgeons to remove the surgical lights.
-
2.
Compose the brain scene viewed by the HS snapshot camera so that the HS linescan camera will be able to capture the entire desired scene. This is accomplished by setting the appropriate height and tilt angle, as illustrated on the left of Fig. 4(a), with the arrows next to the motorized linear stage and sensors. Then, fix the system position and do not move it unless necessary.
-
3.
Focus the HS snapshot camera with the servomotor controlled with the GUI developed from custom C++ software.
-
4.
Capture a RGB image for reference with the RGB sensor included in the depth camera. Then, measure the working distance and tilt angle from the cameras to the area of interest. Every time the distance is measured, depth and RGB .png images are automatically saved with the brain scene, as presented in Fig. 4(a) on the right side below the depth camera.
-
5.
Capture and save HS snapshot images at 60, 70 and 80 ms of exposure time, generating three .tif raw images. One of these captures is illustrated at the centre of Fig. 4(a) below the HS snapshot camera. Note that images from patients with ID numbers of 177 or higher are taken at 90, 100 and 110 ms of exposure time.
-
6.
Record and save a video of around 10 s with the HS snapshot camera with frames taken at a 70 ms exposure time and a frame period of 80 ms. At the same time, record RGB and depth map videos using the depth camera with the same duration. The frame rate of the depth camera is set to 30 FPS. The RGB video with the RGB sensor included in the depth camera has dimensions of 1920 × 1080 pixels, whereas the depth video has dimensions of 1024 × 768 pixels. Step 6 produces three different .bin video files, as shown in Fig. 4(a), below the three images with the .bin icon, which are a raw video for the HS snapshot camera and two videos with RGB and depth information captured with the depth camera. When the Azure Kinect depth camera was used (for patients with ID numbers of 177 or higher), the RGB video had 3840 × 2160 pixels, whereas the depth video had 640 × 576 pixels.
-
7.
Proceed with a multiview scan. This scan uses a depth camera to capture RGB images and depth maps as well as HS snapshot images. As indicated by the arrow pointing left below the system in Fig. 4(b), all three kinds of images are captured while moving the motorized linear stage in one direction. The system first stops and then obtains all 3 captures. Then, it moves 1 cm to capture again until the system stops to capture at least 7 times. Therefore, during the multiview scan, at least 7 .tif raw images are obtained with the HS snapshot camera, as are 7 or more depth and RGB .png images. All these images can be seen on the right of Fig. 4(b), indicating the image format produced by each of the sensors and a timeline below representing that the images are taken sequentially over time. Note that no multiview scans are available for patients with ID numbers of 177 or higher.
-
8.
Proceed with the HS linescan in the other motor direction to compose the brain scene, capturing 800 spatial lines. The left side of Fig. 4(c) shows the scanning direction of the HS linescan camera with the arrow pointing right. Lines are captured with an exposure time of 60 ms and a frame period of 100 ms. This results in an HS cube with spatial dimensions of 1600 × 800 pixels and 394 spectral bands. Later, those bands are trimmed to obtain the 369 effective spectral bands specified by the manufacturer. This linescan procedure generates a raw binary file with all spatial and spectral information for every line as well as a header file. The latter specifies how the data need to be read to compose the HS cube. The right side of Fig. 4(c) illustrates the timeline in which HS lines are taken sequentially to conform to the brain scene on the right, which is saved with the two mentioned files indicated with the .bin and .hdr icons. Note that the exposure time, frame period and captured lines are set to 150 ms, 160 ms and 500 lines, respectively, for patients with ID numbers equal to or greater than 177.
-
9.
Move the acquisition system away from the patient so that neurosurgeons can continue with the surgery and leave the room.
Schematic of the data acquisition procedure followed in the operating room with the simplified 3D model of the SLIMBRAIN prototype version 3. The red outlines indicate that the sensor was used during the mentioned steps. The images on the right illustrate the files generated during the steps performed on the left with the acquisition system.
If appropriate, the neurosurgeon often allowed the researchers to enter the room to follow the same procedure once the tumour is resected from the brain. This resulted in capture immediately after craniotomy and sometimes also in capture after resection for the same patient. The acquisition procedure was cancelled when there was a problem with the patient that required neurosurgeons to continue with the intervention.
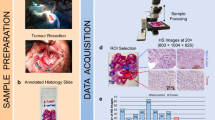
Step 3. Neuropathological assessment of resected tissue
After a piece of brain tissue is removed during surgery, it is sent to the neuropathology laboratory. The tissue is then preserved with formalin and stained with haematoxylin and eosin as well as any other necessary staining techniques to make a definitive histological diagnosis. In commonly used clinical practices, neuropathologists differentiate between tumourous and normal brain samples and provide a histological diagnosis. Additionally, tumour samples are classified based on their histopathological diagnosis, type, and grade.
Step 4. Data preprocessing
Hyperspectral images taken with the snapshot and linescan cameras were preprocessed following a similar procedure. The preprocessing chain used consists of the following steps: hyperspectral cube formation, spectral band removal (only for the linescan camera), data calibration, spectral correction (only for the snapshot camera) and pixel normalization. Additionally, captures were cropped and rotated when necessary to help neurosurgeons during the data labelling procedure. This was important for them to have captures similar to what they were seeing during surgery. Furthermore, all the HS preprocessed data are stored in MATLAB binary (.mat) files, which store different variables for some of the data preprocesing steps. The detailed steps are described below.
Data calibration and calibration libraries
The data captured with the HS camera sensors gather uncalibrated raw information or digital numbers of samples without any meaningful physical units. Thus, to obtain the reflectance of a sample, it is necessary to eliminate the effects of the HS sensor and the lighting conditions captured with the raw images. The reflectance of the brain surface is calculated by capturing dark and white reference images under the same conditions as those used for brain capture. Because the dark reference is obtained by covering the HS camera lens with its lens cap, the tilt angle, distance, and light source used do not matter when the image is taken. However, the white reference must be taken at the same tilt angle and distance and use the same light source conditions as used for the capture. The white reference is obtained by capturing a Lambertian diffuse target with 95% reflectance values (SphereOptics GmbH, Herrsching am Ammersee, BY, Germany). Once all the captures are taken, Equation (1) is used for each spectral band to obtain the calibrated and reflectance cube Ic. Here, I is the brain capture, D is the dark reference and W is the white reference.
First, white and dark references were taken right after the raw brain image in the operating theatre. However, multiple calibration libraries have been captured to ease the calibration process and avoid having to capture a target reference image after the brain image is captured during the intervention. These calibration libraries were generated in the same operating room when it was not in use. Furthermore, white references have been taken by placing the diffuse target reference in the same operating bed and taking images at different working distances and tilt angles. The acquisition of white references is presented in Fig. 5(a), where the acquisition system is tilted to obtain white references of the diffuse reflectance target. Here, when the angle = 0 the cameras point to the x-axis. The more tilted the cameras are, the closer the system points to the z-axis. In Fig. 5(b), multiple white and dark references are shown for both HS cameras. In these calibration libraries, all possible capture combinations were taken at distances of approximately 5 cm and 10 degrees from each other. Due to these differences between images, a rounding function is used when measuring the distance and tilt angle of every brain image taken. In this way, for every raw brain image the closest white reference is utilized to calibrate the scene. All these dark and white references are presented in Table 2 and will be described in further detail in the Data Records section.
Schematic of the calibration library procedure followed in the operating room with the simplified 3D model of the SLIMBRAIN prototype 3. (a) Illustration of the positioning of the acquisition system with respect to the diffuse reflectance target. (b) White and dark references obtained with the Ximea snapshot and Headwall linescan cameras.
Hyperspectral cube formation and spectral band removal
The sensor included in the HS snapshot camera from Ximea provides a 2D image with 2045 × 1085 pixels in repeating mosaic blocks of 5 × 5 pixels. These mosaics contain all 25 spectral bands for the same spatial pixel. Therefore, the 2D image must be rearranged into a 3D cube. The final data dimensions are 409 × 217 pixels in spatial resolution with 25 spectral bands. The cube formation is illustrated in Fig. 6(a), which shows how the spectral information is stored in the 2D HS image inside the mosaics and how, by arranging all of them, the 3D HS cube is obtained.
(a) Representation of the mosaic pattern included in the sensor of the Ximea snapshot HS camera. The 2D image contains the spatial dimensions as well as the spectral information, where the 5 × 5 mosaic contains all captured wavelengths for the same spatial pixel. Therefore, a cube arrangement is performed to obtain a 3D HS cube with 5 times less spatial resolution than that captured in the 2D HS image. (b) Reconstructed HS snapshot cube with a representation of how it is saved in BSQ format.
Note that the spatial band dimensions from the 3D HS cube are 5 times lower than those from the 2D HS image. However, not all captures retained the same spatial dimensions; they needed to be cropped to display only the relevant region of interest to neurosurgeons in the labelling process. Furthermore, not all captures have been saved as raw .tif images but as binary data with a header using the ENVI format. The header file is necessary to determine how the binary data need to be interpreted properly. In particular, binary data are stored in band sequential (BSQ), which saves all rows of a spectral band before saving the information of the next band. The BSQ scheme is illustrated in Fig. 6(b), which shows how the hyperspectral information from a reconstructed cube is saved in binary form with a header file.
Conversely, the HS linescan camera from Headwall captures all the spectral bands for all the pixels in a single spatial line. Hence, once the camera is triggered, it captures a line with 1600 pixels and all 394 spectral bands. Once the camera scanning procedure is complete, it is saved as a binary file with a header following the ENVI format. The binary file stores the information in band-interleaved-by-line (BIL) format, meaning that it saves all the bands of a spatial line before saving the information from the next line. Then, the header specifies how the data should be read to arrange all the captured lines into a hyperspectral cube. This procedure is illustrated in Fig. 7. The image on the left shows the information that each line contains and how a scanning procedure is needed to obtain the desired scene. In the centre, the image presents how the information of each line is stored in memory following the BIL format. Finally, the image on the right shows how, by reading the header file of the binary data, a 3D HS cube is arranged.
After a cube with all the lines captured by the linescan camera is arranged, only the effective spectral bands between 400 and 1000 nm are retained; the rest are removed. This produces hyperspectral cubes with 1600 × Mlines × 369 dimensions, with Mlines = 800 for all the captures obtained with the SLIMBRAIN prototype version 3. In addition, the HS linescan cubes captured with the SLIMBRAIN prototype version 4 have Mlines = 500, as mentioned in step 8 of the Data acquisition subsection. Nonetheless, not all cubes maintain the same spatial dimensions, as they are cropped to display the relevant region of interest to the neurosurgeon, as already indicated with the Ximea snapshot cubes.
Spectral correction
The mosaic sensor of the HS Ximea snapshot camera generation 1 has two issues. First, some filter response curves determined during sensor production exhibit secondary harmonics that are eliminated with a Thorlabs FELH0650 longpass filter (650 nm cut-off wavelength). Second, the response curves also show crosstalk between adjacent pixels, which varies with the angle of incident light to the sensor. This variation is due to the different cavity lengths of the Fabry–Pérot filters in the 5 × 5 mosaic and occurs at the maximum wavelength of neighbouring pixels. To correct this effect, any HS cube must be spectrally corrected after calibration by multiplying it by a sensor-specific correction matrix provided by IMEC. Using Equation (2), any calibrated HS cube Ic can be multiplied by the spectral correction matrix SCM to obtain a spectrally corrected HS cube Isc. Note that Ic has only been calibrated using Equation (1).
Cube cropping and rotation
Ximea snapshot and Headwall linescan images are always cropped. This is done to help the neurosurgeon perform the labelling procedure to focus only on the region of interest while removing the pixels from the draping material around the patient. Therefore, the preprocessed hyperspectral cubes do not retain the maximum spatial dimensions obtained with the cameras (409 × 217 pixels for the snapshot camera and 1600 × 800 pixels for the linescan camera). Hence, the spatial dimensions between the hyperspectral cubes vary depending on how the image was taken. Furthermore, the Headwall linescan captures are rotated 90 degrees counterclockwise, as the camera is attached to the right side of the SLIMBRAIN prototype version 3. However, in version 4, the HS linescan camera position causes vertical flipping of the captures, which is solved when processing the data. The adjustments of the captures were requested by the neurosurgeons to be able to situate the brain in relation to how they saw it in the operating room and how the Ximea snapshot image was taken.
Pixel normalization
Pixel normalization is carried out for each capture to compare the spectral signatures obtained from the captures made under different lighting situations. In normalization, the root mean square (RMS) of a pixel energy is calculated over all bands and is used as a normalizing coefficient (Equation (3)), which normalizes the spectral corrected cube as expressed in Equation (4), where Isc is the spectrally corrected cube with dimensions r × c × b (rows × columns × bands). Note that with the files included in the SLIMBRAIN database, any pixel normalization can be applied, as the calibrated-only HS cubes are provided.
Step 5. Ground-truth labelling procedure
Once the hyperspectral cubes have been preprocessed after surgery, pseudo-RGB images of the in vivo brain images are created by selecting the 3 most appropriate wavelengths out of all the captured images. The synthetic RGB images are generated using Ximea snapshot wavelengths of 845.01 nm (red), 740.65 nm (green), and 670.68 nm (blue), whereas the Headwall linescan VNIR camera uses 709.35 nm (red), 540.39 nm (green), and 480.27 nm (blue) wavelengths for its channels. These pseudo-RGB images are necessary to conduct the ground-truth (GT) labelling procedure to generate the GT maps presented in Fig. 8 and are discussed in further detail later in this subsection. The obtained GT maps are stored in MATLAB binary (.mat) files to include relevant data regarding the labelled pixels. However, it is not possible to obtain a complete GT of the brain capture because living humans are involved. Obtaining a complete reference GT would require the pathologist to analyse the entire brain tissue exposed, which is impossible for ethical reasons, as it would require the neurosurgeon to cut all the tissue, which would pose grave risks to the health of the patient. Therefore, to obtain partial GT maps, we rely on the experience and knowledge of the operating neurosurgeons and the pathological analysis of a tumour sample to locate its position in the capture as well as the pathology itself. With both criteria, the neurosurgeon is presented with an interactive graphical user interface designed in the MATLAB GUIDE application (The MathWorks Inc., Natick, MA, USA) to label the pixels of interest in the capture. Although the inspection of the brain scene is performed visually by the neurosurgeon, the labelling tool is based on the spectral angle mapper (SAM) algorithm37 to reduce errors when obtaining the partial GT map. Then, the neurosurgeon labels healthy, tumour, venous, artery, and meningeal pixels on the synthetic RGB as accurately as possible.
On the left side of Fig. 8, step 1 presents the preprocessed HS cube overlaid with the pseudo-RGB for the neurosurgeon to select a reference pixel of the tissue to label. Steps 2, 3, and 4 are shown in the centre of Fig. 8. In step 2, the SAM algorithm detects pixels in the HS cube with similar spectral angles as the reference pixel by manually varying a threshold. Then, a binary mask is generated and used to highlight all the pixels with a spectral angle lower than the defined threshold. Once the threshold is fixed to identify coincident physiological features of the chosen tissue, the neurosurgeon selects the desired pixels with a region of interest in step 3 and provides them with a label in step 4. Instead of choosing from a larger variety of questionable pixels, neurosurgeons were advised to choose only a small number of groupings of very dependable pixels. These four steps are repeated until a complete GT map with the desired labelled tissues is generated, as depicted by the timeline on the right side of Fig. 8. This labelling tool has already been used in previous in vivo hyperspectral human brain image databases26, and as already indicated in previous studies, two key benefits in creating GT maps can be noted. First, the pseudo-RGB masked image, which displays the pixels with lower spectral angles than the reference pixel, can be used to confirm that the reference pixel chosen by the expert does, in fact, belong to a particular class. Second, manually choosing pixels from an HS cube for each class is a laborious operation; thus, this semiautomatic approach makes it possible to produce the GT quickly. In the first step, the pseudo-RGB is presented to the neurosurgeon to select a reference pixel. Then, a black image with the same dimensions is displayed on its right, showing only the selected reference pixel. Later, the neurosurgeon adjusts a threshold to show similar pixels in the masked black image. This uses all the spectral information from the preprocessed HS cube to find similar pixels based on the SAM metric. Once the neurosurgeon thinks that enough relevant pixels are displayed, a mask is created around the pixels to tag them with a predefined label. Notably, that the GTs were completed days after the intervention, thereby affording neurosurgeons access to pertinent patient data, including magnetic resonance images and medical treatment, for the purpose of accurately categorizing the hyperspectral images acquired. The categories utilized are straightforward and readily discernible by neurosurgeons. The differentiation between meninx, artery, and vein is unambiguous, as there is no ambiguity regarding their identification. However, distinguishing between tumour and healthy tissue remains the most challenging aspect. The labelling criterion that we have followed is that the observer for this task is the neurosurgeon who performed the intervention, as they are the most familiar with the patient to identify the location of the lesion and interpret the imaging data. The labelling tool used by all experienced neurosurgeons is semiautomated and generates an objective measure based on the SAM to identify pixels that are spectrally similar to a reference pixel. Although the tool does not eliminate the influence of the neurosurgeon, it attempts to standardize the procedure across different observers to label the data. Furthermore, the reference pixels for the tumour tissue are selected based on the MRIs available, allowing for more reliable identification of the tumour. In contrast, the healthy tissue pixels are labelled as far away from the tumour as possible. Because the reliability of each individual neurosurgeon is not known, given the complexity of the process for obtaining these values, the reliability of the GT obtained is likely to be affected. Although this structured procedure for labelling helps reduce the number of errors made by neurosurgeons, there is a lack of intra- or interrater reliability of ground truth data, as only one neurosurgeon labels the GTs.
Step 6. Patient data extraction
To extract labelled patient data, the GT map and its corresponding preprocessed HS cube are needed. Fig. 9 illustrates how the coordinates of the labelled pixels in the GT on the left are extracted and used to gather the normalized reflectance values from the preprocessed HS cube at the centre. Mapping the GT pixels to the HS cube generates a .mat file containing a 2D matrix of reflectance values and a 1D vector with the corresponding labels. The previous matrix and vector are presented on the right of Fig. 9, which can be accessed using the data and label keys in the generated IDxCy_dataset.mat file for patient x and capture y. Note that the labelled patient data are extracted for each patient independently.
Data extraction of labelled pixels performed for every patient capture. This process uses the GT map to obtain the coordinates on the preprocessed cube to obtain the reflectance information. This procedure generates a .mat file including a matrix with all spectra reflectances as well as a vector with the label provided for every pixel. Each image in the figure has a key located above it, which is used to access the information in the .mat file to present its image below. Furthermore, examples of the .mat files used are presented below each of the three images (i.e., SNAPgtID000067C01.mat contains the ‘groundTruthMap’ key with the information to represent the GT map).
Data Records
The data records cited in this work are available from the e-cienciaDatos repository38. E-cienciaDatos is a multidisciplinary data repository that hosts the scientific datasets of researchers from the public universities of the Community of Madrid and UNED, members of the Madroño Consortium, to make these data visible, guarantee their preservation and facilitate their access and reuse. The data available in the repository consist of 6 zipped folders, which are described throughout this section. Importantly, lamp A from Fig. 3 was used from patient ID000001 to ID000135 and lamp B from the same figure was used from patient ID000136 to ID000176. All these images were captured with the operating theatre fluorescent lights (lamp D) turned on. Lamp C from Fig. 3 was used from patient ID000177 onwards, as it provides more energy in the NIR spectra than the previous lamps did. Although SLIMBRAIN is an expanding database, with patient data being uploaded on a weekly basis, the information detailed in this manuscript pertains to a subset of the database that corresponds to version 4.
Raw data and calibration files
Table 1 summarizes all the raw files obtained during the data acquisition procedure with all the versions of the developed acquisition system. These files can be found inside the zipped folder “RawFiles” on the e-cienciaDatos repository, as well as in the RawFiles directory. All the files are described with /RawFiles/IDx, where x is a six-digit number used to identify each patient. First, the System column indicates the equipment used to generate each raw file, including the three HS cameras (Ximea snapshot, Ximea linescan and Headwall linescan), a mobile phone, the PC managing all the sensors and actuators and the depth cameras (Intel LiDAR L515 and Azure Kinect). Second, the Subdirectory column indicates the name of the subdirectories that store different types of raw files below the parent directory previously mentioned. This information is included for all systems in the first column. The subdirectory names include two numbers, x and y, where x a six-digit number that identifies each patient and y is a two-digit number indicating the capture taken for patient x. The difference among subdirectory names for the same system depends on the patient ID. For example, the Ximea snapshot system subdirectories are different from patient ID 70, as presented in the first two rows of Table 1. Patients IDs 1 to 70 have the /images_IDx subdirectory for raw hyperspectral images, whereas patient IDs equal to or greater than 71 have the /ID_x_CN_y subdirectory name. This x and y nomenclature is used throughout the table. Note that for every patient x, multiple captures can be obtained. For example, one capture could be obtained after the craniotomy and durotomy, where the brain surface is exposed, and another capture could be obtained when part of the tissue has been resected. Third, the File name column provides the different names that the raw files can have. Here, the “*” punctuation mark represents a wildcard indicating that the file can have any suffix. Additionally, for a sequence of consecutively taken images, z denotates the order in which every image was taken. Multiple file formats can be found for the same file name. This is notated with curly brackets, expressing that a file name can have one or multiple formats. Fourth, the Data type column expresses the type of data stored for each of the raw files. Fifth, the Patient IDs column indicates to which patient the data in the Subdirectory, File name and Data type columns apply. Finally, the sixth column provides a general description of the raw file. The raw files used to calibrate the hyperspectral images are described in Table 2, which can be found inside the zipped folder “CalibrationFiles” on the e-cienciaDatos repository, as well as in the CalibrationFiles directory. First, the Systems column indicates the collection of capture systems used to generate each of the five calibration libraries. Second, the Lamp column indicates which light lamp (the spectral responses of which are presented in Fig. 3) has been used to illuminate the diffuse target with 95% reflectance to obtain white reference images. Third, the Subdirectory column notates the subdirectory to find the calibration library in the parent directory, which is /CalibrationFiles. Fourth, the Camera column expresses which camera an HS images can be calibrated with the white references in each calibration library. Fifth, the White reference file name column indicates the nomenclature used in each calibration library to describe the different white references. Here, X and Y are the distance in centimetres and the angle in degrees, respectively, at which the white reference has been captured. The Z letter expresses the time exposure at which the capture has been taken, where Z = 1, Z = 2 and Z = 3 indicate 60 ms, 70 ms and 80 ms, respectively. For some Ximea snapshot white references, the letter B notates the bit depth used, where B = 8 and B = 16 are captures of 8 and 10 bit depths, respectively.
Moreover, the exptime also expresses the exposure time in milliseconds at which some Ximea snapshot white references have been captured. Although exptime describes the exposure time in milliseconds with five digit numbers, Z only uses one digit number (either 1, 2 or 3, as previously described). Finally, the Patients column indicates which patient captures have been calibrated with each calibration library and camera.
Furthermore, the intrinsic parameters for the Intel RealSense L515, Azure Kinect and HS snapshot cameras are provided within the /CalibrationFiles/DepthCameraCalibrationFiles/ directory. Inside, two .json files can be found, cam_params_Intel_L515_Ximea_Snapshot.json and cam_params_Azure_Kinect_Ximea_Snapshot.json. Both .json files include not only the intrinsic parameters for each image sensor but also the extrinsic parameters between the sensors described in each file. For example, cam_params_Intel_L515_Ximea_Snapshot.json includes the extrinsic parameters between Intel L515 infrared, RGB and Ximea HS snapshot sensors, and the cam_params_Azure_Kinect_Ximea_Snapshot.json includes the extrinsic parameters but for the Azure Kinnect depth and RGB sensors and the Ximea HS snapshot sensor. The following values are included in each file: camera rotation and translation, distortion, intrinsic matrix and sensor resolution in pixels. Notably, the coordinate system origin is centred on the infrared (IR) sensor in both cases.
Preprocessed hyperspectral data
All preprocessed hyperspectral data from both cameras can be found inside the zipped folder “preProcessedImages” on the e-cienciaDatos repository, as well as in the preProcessedImages directory. In the latter case, the parent directory for the Ximea snapshot preprocessed HS data is /preProcessedImages/Ximea_Snapshot, whereas the preprocessed data of the Headwall linescan camera are located at /preProcessedImages/Headwall_Linescan. Table 3 details the preprocessed HS data records. First, the Camera column indicates which camera the preprocessed data are coming from. Second, the File name column notates the nomenclature employed to represent each preprocessed file, with x representing a six-digit number to identify the patient and y representing a two-digit number to identify the capture number. The “*” punctuation mark is a wildcard that indicates that the file suffix is “_cropped_Pre-processed”. Third, the Spectrum (nm) column presents the spectral range measured with each camera and is saved in their corresponding preprocessed .mat files. Fourth, the Bands column shows the number of spectral bands measured within the spectral range from the Spectrum (nm) column. Fifth, the Key column presents the keys available in the preprocessed .mat files to access different stored information. Finally, each key is described in the Description column.
Ground-truth maps
All ground-truth maps from both cameras can be found inside the zipped folder “GroundTruthMaps” on the e-cienciaDatos repository, as well as in the GroundTruthMaps directory. In the latter case, the parent directory for the Ximea snapshot labelled HS cubes is /GroundTruthMaps/Ximea_Snapshot, whereas the labelled GT of the Headwall linescan camera an HS cubes is /GroundTruthMaps/Headwall_Linescan. These GT map files are described in Table 4, and all possible labels included in the Ximea snapshot and Headwall linescan GT maps are described in Table 5. First, in Table 4, the Camera column indicates the camera used to obtain the labelled HS preprocessed cube. Second, the File name column notates the nomenclature employed to represent each GT map file, with x representing a six-digit number to identify the patient and y representing a two-digit number to identify the number of the capture. The “*” punctuation mark is a wildcard that indicates that the file suffix is “_cropped_Pre-processed”. Third, the Key column presents the keys available in the GT .mat files to access different stored information. Finally, each key is described in the Description column. Additionally, in Table 5, the first column, named Label, is a three-digit number used to differentiate tissues. Second, the Label class column provides a generic tissue description for each label. Third, the Tissue column provides a detailed description of the tissue label. Finally, the Pixels and Pixel (%) columns represent the number of labelled pixels and their total percentage for the corresponding tissue label for every camera.
Table 5 includes 145 GT maps from patient captures from ID000017C05–ID000153C02, for a total of 680,911 labelled pixels. It also includes 17 GT maps from patient captures obtained with the HS linescan camera from ID000120C01-ID000149C01, summing a total of 1,039,147 labelled pixels. These 145 GT maps used from the HS snapshot camera can be found inside the zipped folder “PaperExperiments” on the e-cienciaDatos repository, as well as in the Ximea_Snapshot_GT_Patient_IDs.txt file. Furthermore, the 17 GT maps used from the HS linescan camera can also be found inside the zipped folder “PaperExperiments” on the e-cienciaDatos repository, as well as in the Headwall_Linescan_GT_Patient_IDs.txt file.
Patients labelled data
All patients labelled patient data from both cameras can be found inside the zipped folder “Datasets” on the e-cienciaDatos repository, as well as in the Datasets directory. In the latter case, the parent directory for the Ximea snapshot labelled data are /Datasets/Ximea_Snapshot, whereas the labelled GT maps of the Headwall linescan camera an HS cubes is /Datasets/Headwall_Linescan. Table 6 details each patient data record containing the reflectance values for every labelled pixel. First, the Camera column indicates the camera used to obtain the HS preprocessed cube from which each patient dataset has been obtained. Second, the File name column notates the nomenclature employed to represent each GT map file, with x representing a six-digit number to identify the patient and y representing a two-digit number to identify the number of the capture. Third, the Key column presents the keys available in the each patient dataset .mat file to access different stored information. Finally, each key is described in the Description column.
Technical Validation
Although three HS cameras have been utilized to acquire images, the images taken from the Ximea linescan camera presented many difficulties when reconstructing HS cubes with the nonflat surface of the brain. These issues have been described in published works39, with the most noticeable being that the black stripes, with empty information, remain in the postprocessed reconstructed HS cubes because the actuator that holds the camera is not moving precisely. Therefore, the corresponding hyperspectral cubes have not been reconstructed properly to be labelled. Despite this fact, the raw captures of this camera have been included in the SLIMBRAIN database for any researcher to conduct further experiments.
Throughout this section, multiple validations are conducted to show the quality of the data. First, the spectral signatures obtained with the HS cameras over a polymer reference are provided, which are helpful for determining the quality of the cameras when capturing images in the operating room. They are compared with the spectral response of the polymer provided by the manufacturer as well as the response measured with a spectrometer. Furthermore, the spectral signatures from the labelled in vivo brain tissues are presented for both HS cameras, indicating the similarity among tissues and any strange behaviour noted, such as the influence of lamp D (ambient fluorescence). Second, the GT maps are validated by measuring the SAM threshold set for each reference pixel used to label similar tissues. Third, the depth images obtained are validated by performing state of the art virtual view generation analysis. These images are necessary to ensure that the distance measured by the depth cameras is correct to precisely superimpose the HS image over a generated 3D brain scene with multiview or depth images. Finally, an ML classification experiment is performed on data from both cameras to predict a test set, which provides information regarding the usability of the data to classify brain tissue.
Spectral signature validation
Figure 10 presents evidence to validate the spectral signatures obtained with the HS cameras and the spectrometer in the operating theatre. The x- and y-coordinate axes represent the wavelengths and the normalized reflectances, respectively.
Normalized reflectance spectral responses with correlation values when illuminating the polymer reference with lamps B and D from Fig. 3 and captured with different sensors. Spectrometer measurements were performed with a fibre optic orthogonal to the polymer reference from a 5 cm distance. The hyperspectral camera captures a scene of the polymer at approximately 72.5 degrees and 40.5 cm distance and uses a region of interest of 25 × 25 pixels. Note that captures taken at 90∘ and 0∘ would mean pointing the camera to a vertical wall and to the floor, respectively. These responses from the HS cameras are the mean spectral signatures of the 25 × 25 pixel polymer pixels with their corresponding standard deviations, shown with shaded colours between their corresponding means. The spectrometer uses 2055 bands and covers the 350–925 nm range. The HS linescan VNIR camera covers the 400–1000 nm spectral range and uses 369 bands. Finally, the HS snapshot NIR camera spectra ranges from 660–950 nm and includes 25 bands. The Pearson correlation is presented after the measured bands of each sensor are compared with the polymer reference response. The visible (VIS) spectra is illustrated with rainbow shading in the 380–740 nm range.
As shown in Fig. 3, the spectral signature of the fluorescent light in the operating room presents very pronounced peaks in the visible spectra. Therefore, a reference material with a very well-defined spectral response was used to analyse the spectra measured with the mentioned sensors and determine the influence of the fluorescent light. The zenith polymer wavelength standard used as a reference consists of a polymer with nearly ideal Lambertian diffuse reflectance (SphereOptics GmbH, Herrsching am Ammersee, BY, Germany). First, the spectral response of the polymer reference is provided by the manufacturer and is illustrated by the red line in Fig. 10. Second, the spectral response measured with the spectrometer is illustrated by the blue line from 350-925 nm. The Pearson correlation coefficient of the obtained spectral signature with respect to the red reference is 91.72%, when 2055 measured bands captured with the spectrometer are used. If bands from 350–400 nm are removed (as shown in Fig. 10, the information acquired is essentially noise, as there is no energy illuminating this part of the spectra), the Pearson correlation is 96.91% when 1913 bands are used. Additionally, these measurements were obtained by using a fibre optic pointing orthogonal to the illuminated polymer reference at a distance of approximately 5 cm. Third, the mean spectral response of the polymer obtained with the Headwall linescan camera is illustrated by the green line, starting at 400 nm and ending at 1000 nm. The Pearson correlation coefficient with the red line reference is 95.87% when the 369 effective acquired bands are used. Finally, the mean spectral response of the polymer captured with the Ximea snapshot camera is presented from 660–950 nm range with the orange line. The correlation obtained with respect to red line reference is 68.19% when the 25 spectral bands captured are utilized. Even though the correlation is lower than that obtained with the spectrometer or the other HS camera, the orange line clearly illustrates how the spectral response is quite similar from the 660–875 nm. We recomputed the Pearson correlation correlation with 17 bands from 660–866 nm, and the obtained result is indicated a correlation of 95.55%, demonstrating that not enough energy is captured with the latest 8 bands. As seen from 866–950 nm in the orange line in Fig. 10, the mean spectral response of the Ximea snapshot camera decreases progressively and does not behave similarly to the polymer reference in the red range as it does in the 660–866 nm range. Note that the spectrometer, HS linescan and HS snapshot spectral signatures were obtained under illumination with lamps B and D from Fig. 3.
For ease of reading, an in-depth analysis of the in vivo human brain spectral signatures obtained with both cameras is provided in Figs. S1 and S2, which are available in the Supplementary Materials. Furthermore, the influence of ambient light (lamp D) on the spectral signatures obtained is analysed in Figs. S3–S5.
Ground-truth map validation
To validate the GT maps generated with the help of the neurosurgeons, an analysis of the SAM thresholds for every reference pixel was performed. This analysis is illustrated in Figs. 11 and 12, which shows a raincloud plot40, a very useful illustration that addresses the issue of data obscuration when presenting error bars or box plots.
Raincloud plot with the SAM thresholds used for every reference label pixel. The images obtained during the ground-truth labelling procedure for the Ximea snapshot brain capture with IDs can be found inside the zipped folder “PaperExperiments” on the e-cienciaDatos repository, as well as in the Ximea_Snapshot_GT_Patient_IDs.txt file.
These charts combine different data visualizations to display raw data (with single data points), probability density (with half violin plots), and essential summary statistics, including median, first and third quartiles, outliers, and relevant confidence intervals (with boxplots), in a visually appealing and adaptable way with minimal redundancy. Every data point is a single SAM threshold set by neurosurgeons when a reference pixel is selected during the GT labelling, as illustrated in Fig. 8, after performing steps 1 and 2. Note that the data point distribution width is irrelevant but has been added because it helps to see the data points better than in a line. All GTs store the SAM thresholds defined for every reference pixel, regardless of the label selected. Figure 11 shows the thresholds obtained from the 145 GT maps with IDs available, which can be found inside the zipped folder “PaperExperiments” on the e-cienciaDatos repository, as well as in the Ximea_Snapshot_GT_Patient_IDs.txt file. The red dots inside the boxplots indicate the mean value of each distribution, which are connected between tissues to visualize the mean tendency across them. Note that the ganglioma, necrosis, non defined blood and blood clot labels have been removed, and a single reference pixel has been used for each class. In addition, lung metastasis (2 patients), breast metastasis (1 patient) and skin metastasis (4 patients) images were gathered on a single metastasis label. The dura matter and arachnoid labels were also combined into a single meninges label. Although few outliers appear in the raincloud plot, the continuous red line indicates that the mean SAM threshold values used for every labelled pixel are between 20 and 30%. These results indicate that the labelled pixels on almost every GT map use a low threshold when analysing the similarity of the pixels from the reference map, which is shown in the example in Fig. 8, step 3, when an SAM threshold is set and only spatial pixels relevant to the reference pixel are selected. Therefore, fewer pixels may have been labelled in a class to which they do not correspond, providing GT maps with reasonably good labelled pixels. The same analysis was conducted for the 17 available GT maps generated for the Headwall linescan captures.
The SAM threshold analysis is illustrated in Fig. 12, which reveals that the mean SAM threshold values range from 28–37%, as indicated by the continuous red line. Although the mean is higher than that from Fig. 11, remarkably, fewer reference pixels have been used because approximately eight times fewer images have been labelled. Therefore, a more robust value of the SAM mean among tissues could be provided with more GT maps from the Headwall linescan captures. Nonetheless, testimonials from neurosurgeons indicate that the ease of labelling these captures has been more accurate and easier than the Ximea snapshot captures when delimiting more pixels of the same tissue, resulting in a higher threshold to provide greater number of labelled pixels.
Raincloud plot with the SAM thresholds used for every reference label pixel. Obtained during the ground-truth labelling procedure for the Headwall linescan brain captures with IDs, which can be found inside the zipped folder “PaperExperiments” on the e-cienciaDatos repository, as well as in the Headwall_Linescan_GT_Patient_IDs.txt file.
Depth image validation
To validate the depth images captured in the operating room, a virtual view generation analysis was performed. This analysis employs the multiview RGBD (RGB + Depth) captures present in the SLIMBRAIN database plus the Reference View Synthesis software (RVS)41,42, a state of the art video synthesis tool. This methodology allows indirect depth map validation in scenarios where measuring the real distance from the camera to the objects is not possible, as illustrated in Fig. 13(a).
Notably, the quality of the virtual view is completely dependent on the RGBD depth quality. To employ this approach, it is necessary perfrom the multiview geometric calibration, i.e., the relative position between the captured images. This information is known, as the captures are always displaced 1 cm in a single dimension. However, due to system vibrations or unexpected movements during the recording, the calibration may vary. For this reason, the iterative closest point (ICP)43 algorithm is employed to refine the calibration. This algorithm is employed to match all the RGBD captures in the same 3D space, performing affine transformations. In this way, all the RGBDs captured in a scan automatically share the same coordinate system. By recording the transformations performed for each capture to find the match, a new calibration is generated and can be introduced into the view synthesis tool. The analysis is performed by emulating the action performed in a real operation where the LiDAR is employed to generate an immersive video representation from different points of view. To do so, the scheme depicted in Fig. 13(a) is followed. From the multiview array, the central capture (view 2 in the example) is utilized to generate an RGB virtual view in the position of all the other real views (1). Then, they can be objectively compared to the real views in the same positions (2). To perform the virtual view generation, RVS is employed. For the objective comparison, IV-PSNR44 is employed. IV-PSNR is an objective quality metric specifically designed to assess the quality of virtual views in immersive video applications to compare the virtual and real views. For each multiview capture, an IV-PSNR value is generated for each camera view, corresponding to the comparison between a real RGB capture and a virtual RGB synthesis. They are employed to generate an average IV-PSNR that represents the depth quality level for a multiview capture. Figure 13(b) shows the results obtained for all the multiview captures present in the SLIMBRAIN database using a raincloud plot. The results show that the virtual views generated have a median mean IV-PSNR of almost 30 dB and a mean close to 29 dB. To better understand the meaning of these results, a subjective comparison is introduced in Fig. 14. These results show the best-case, mean, and worst-case results in virtual view generation. The best-case and mean virtual views are subjectively equal to the real view, only considering the differences in borders (in the difference image). In the case of the worst IV-PSNR value, the difference is in the position of the camera, showing that the calibration in that capture is not correct. From these results, it can be concluded that it is possible to employ a single RGBD camera to generate virtual views up to 3 cm apart.
Machine learning classification validation
A random forest (RF) model was trained to validate the labelled pixels for classification purposes. RF was selected because it is a well-known state of the art algorithm and is very fast to train when large datasets are used. To avoid bias, three splits were created with different percentages of patient images. These sets are used for training, validation (to perform hyperparameter optimization), and testing, which include approximately 80%, 5%, and 15% of the labelled captures, respectively. Furthermore, captures taken after resection were not included, leading to having only 1 capture per patient. This decision ensures that different captures from the same patient are not in different sets, as that might bias the model. Moreover, the selection of patients for each fold was performed randomly. First, an RF model was trained, validated, and tested with 91, 7, and 9 labelled captures from the Ximea snapshot IDs, respectively. The specific IDs for each set can be found inside the zipped folder “PaperExperiments” on the e-cienciaDatos repository, as well as in the Ximea_Snapshot_GT_Patient_IDs.txt file. A total of 402,376 pixels were used for training, whereas 40,326 and 47,415 pixels were used for validation and testing, respectively. Furthermore, receiver operating characteristic (ROC) curves have been employed to address model performance because of the imbalanced dataset used. In his way, a better evaluation can be performed, as 36.88% of the labelled pixels for training are healthy (grey and white matter), 30.30% are tumour pixels from every labelled pathology, 5.36% are blood pixels from venous and artery vessels, and the remaining 27.46% are meninges pixels from the dura mater and arachnoid. Note that skull bone-labelled pixels have been discarded. The results are included in Fig. 15(a), where the area under the curve (AUC) scores for the healthy, tumour, blood and meninges classes are 77.85%, 56.85%, 70.65% and 80.89%, respectively. Similarly, this experiment was also performed with labelled captures from the HS linescan camera by following the exact same procedure previously described. For this experiment, labelled pixels from 15 HS linescan captures with IDs included inside the zipped folder “PaperExperiments” on the e-cienciaDatos repository, and in the Headwall_Linescan_GT_Patient_IDs.txt file have been employed to train, validate, and test the RF model. Specifically, 12, 1, and 2 labelled captures have been used to train, validate, and test the model, respectively. For each training, validation, and test set there are a total of 670,312, 30,905, and 173,950 labelled pixels, respectively, including all 369 effective bands of the camera. The percentages of pixels in the training set are 54.78% for healthy pixels (grey and white matter), 13.11% for tumour pixels (including all labelled pathologies), 2.95% for blood pixels (venous and artery vessels) and 29.16% of meninges pixels (dura mater and arachnoid). Note that skull bone labelled pixels have been discarded. The results are shown in Fig. 15(b), where the AUC values for the healthy, tumour, blood and meninges classes are 99.06%, 65.48%, 99.31% and 97.38%, respectively. These results show the performance of a simple model when following a simple methodology.
Although other approaches can be followed to increase the performance, it is worth noting that the labelled pixels used come from patients suffering different pathologies, with different biological behaviours, and mix different acquisition systems and different illuminations, all of which probably hinder the model classification. Nonetheless, the results show the potential use of the data to classify in vivo brain tissues. By following other approaches, such as using labelled images captured under the same illumination, using complex ML or deep learning (DL) algorithms, or balancing the data classes for training, studies can expect better classification performance.
Usage Notes
The SLIMBRAIN database is under a data usage agreement (DUA), which users will need to sign prior to accessing the data. The DUA is available to download from the e-cienciaDatos repository38 as well as on the SLIMBRAIN database website. Once the document has been signed, please forward it to all the email addresses listed at the end of the DUA with the subject line “SLIMBRAIN access request”. The providers will evaluate each request and grant access to applicants who meet the specified requirements in the DUA. The criteria for granting access to the data are as follows:
-
The data will be used exclusively for research purposes within the scope of their specified research project, as outlined in Section 6 of the DUA.
-
The users agree to comply with the obligations detailed in Section 5 of the DUA and to adhere to all the IT security principles set forth in Annex 1.
If the data recipient indicates that the data will be used exclusively for research purposes and agrees to comply with the relevant clauses by signing the DUA, they will be granted access in accordance with the terms of the DUA. Then, credentials will be provided to allow access to the e-cienciaDatos data repository38 or search tools on the SLIMBRAIN database website.
Furthermore, guided videos are available on the SLIMBRAIN database website to illustrate how to navigate through the database and use it appropriately. Among these videos, some describe how to navigate through the data records table with detailed and anonymized information of each patient as well as how to visualize RGB brain images.
Code availability
Data included in the SLIMBRAIN database can be opened with a Python package accessible from the SLIMBRAIN database website. This Python software is made by the authors to read any generated .mat file from HS data, including the manipulation of preprocessed HS cubes, GT maps and patient datasets. The package also provides helpful tools to generate classification maps and spectral signature visualizations, among others.
References
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Sato, A. et al. Cerebellar development transcriptome database (cdt-db): profiling of spatio-temporal gene expression during the postnatal development of mouse cerebellum. Neural Networks 21, 1056–1069 (2008).
Shimogori, T. et al. Digital gene atlas of neonate common marmoset brain. Neuroscience research 128, 1–13 (2018).
Tripathy, S., Savitskaya, J., Gerkin, R. & Urban, N. Neuroelectro: a database describing the electrophysiology properties of different neuron types. In Front. Neuroinform. Conference Abstract: 5th INCF Congress of Neuroinformatics. https://doi.org/10.3389/conf.fninf, vol. 99 (2014).
Ascoli, G. A., Donohue, D. E. & Halavi, M. Neuromorpho. org: a central resource for neuronal morphologies. Journal of Neuroscience 27, 9247–9251 (2007).
Jack Jr, C. R. et al. The alzheimer’s disease neuroimaging initiative (adni): Mri methods. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine 27, 685–691 (2008).
Kempton, M. J., Geddes, J. R., Ettinger, U., Williams, S. C. & Grasby, P. M. Meta-analysis, database, and meta-regression of 98 structural imaging studies in bipolar disorder. Archives of general psychiatry 65, 1017–1032 (2008).
Serag, A. et al. Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. Neuroimage 59, 2255–2265 (2012).
Clark, K. et al. The cancer imaging archive (tcia): maintaining and operating a public information repository. Journal of digital imaging 26, 1045–1057 (2013).
Wang, L. et al. Schizconnect: Mediating neuroimaging databases on schizophrenia and related disorders for large-scale integration. NeuroImage 124, 1155–1167, https://doi.org/10.1016/j.neuroimage.2015.06.065 (2016).
Labus, J. S. et al. Pain and interoception imaging network (pain): A multimodal, multisite, brain-imaging repository for chronic somatic and visceral pain disorders. Neuroimage 124, 1232–1237 (2016).
Niso, G. et al. Omega: the open meg archive. Neuroimage 124, 1182–1187 (2016).
Vaccarino, A. L. et al. Brain-code: A secure neuroinformatics platform for management, federation, sharing and analysis of multi-dimensional neuroscience data. Frontiers in Neuroinformatics12, https://doi.org/10.3389/fninf.2018.00028 (2018).
Thakkar, J. P. et al. Epidemiologic and molecular prognostic review of glioblastomagbm epidemiology and biomarkers. Cancer epidemiology, biomarkers & prevention 23, 1985–1996 (2014).
Sanai, N., Polley, M.-Y., McDermott, M. W., Parsa, A. T. & Berger, M. S. An extent of resection threshold for newly diagnosed glioblastomas. Journal of neurosurgery 115, 3–8 (2011).
Miga, M. I. et al. Clinical evaluation of a model-updated image-guidance approach to brain shift compensation: experience in 16 cases. International journal of computer assisted radiology and surgery 11, 1467–1474 (2016).
Orringer, D. A., Golby, A. & Jolesz, F. Neuronavigation in the surgical management of brain tumours: current and future trends. Expert review of medical devices 9, 491–500 (2012).
Gandhe, R. U. & Bhave, C. P. Intraoperative magnetic resonance imaging for neurosurgery–an anaesthesiologist’s challenge. Indian Journal of Anaesthesia 62, 411 (2018).
Albayrak, B., Samdani, A. & Black, P. Intra-operative magnetic resonance imaging in neurosurgery. Acta neurochirurgica 146, 543–557 (2004).
Sastry, R. et al. Applications of ultrasound in the resection of brain tumours. Journal of Neuroimaging 27, 5–15 (2017).
Selbekk, T. et al. Ultrasound imaging in neurosurgery: approaches to minimize surgically induced image artefacts for improved resection control. Acta neurochirurgica 155, 973–980 (2013).
Ferraro, N. et al. The role of 5-aminolevulinic acid in brain tumour surgery: a systematic review. Neurosurgical review 39, 545–555 (2016).
Chung, I. W. H. & Eljamel, S. Risk factors for developing oral 5-aminolevulenic acid-induced side effects in patients undergoing fluorescence guided resection. Photodiagnosis and photodynamic therapy 10, 362–367 (2013).
Lu, G. & Fei, B. Medical hyperspectral imaging: a review. Journal of biomedical optics 19, 010901 (2014).
Fabelo, H. et al. An intraoperative visualization system using hyperspectral imaging to aid in brain tumour delineation. Sensors 18, 430 (2018).
Fabelo, H. et al. in vivo hyperspectral human brain image database for brain cancer detection. IEEE Access 7, 39098–39116, https://doi.org/10.1109/ACCESS.2019.2904788 (2019).
Fabelo, H. et al. Spatio-spectral classification of hyperspectral images for brain cancer detection during surgical operations. PLoS One 13, e0193721 (2018).
Sancho, J. et al. Slimbrain: Augmented reality real-time acquisition and processing system for hyperspectral classification mapping with depth information for in vivo surgical procedures. Journal of Systems Architecture 102893, https://doi.org/10.1016/j.sysarc.2023.102893 (2023).
Martín-Pérez, A., de Ternero, A. M., Lagares, A., Juarez, E. & Sanz, C. Spectral analysis comparison of pushbroom and snapshot hyperspectral cameras for in vivo brain tissues and chromophore identification. Journal of Biomedical Optics 29, 093510, https://doi.org/10.1117/1.JBO.29.9.093510 (2024).
Manuel Villa, G. V. E. A., Alberto, Martín-Perez Libra: Low spectral resolution brain tumour classifier for medical hyperspectral imaging https://doi.org/10.21203/rs.3.rs-4668541/v1 (2024). Preprint on webpage at https://www.researchsquare.com/article/rs-4668541/v1.
Urbanos, G. et al. Supervised machine learning method and hyperspectral imaging techniques jointly applied for brain cancer classification. Sensors 21, 3827 (2021).
Cebrián, P. L. et al. Deep recurrent neural network performing spectral recurrence on hyperspectral images for brain tissue classification. In Chavarrías, M. & Rodríguez, A. (eds.) Design and Architecture for Signal and Image Processing, 15–27 (Springer Nature Switzerland, Cham, 2023).
Martín-Pérez, A. et al. Hyperparameter optimization for brain tumour classification with hyperspectral images. In 2022 25th Euromicro Conference on Digital System Design (DSD), 835–842, https://doi.org/10.1109/DSD57027.2022.00117 (2022).
Vazquez, G. et al. Brain blood vessel segmentation in hyperspectral images through linear operators. In International Workshop on Design and Architecture for Signal and Image Processing, 28–39 (Springer, 2023).
Vazquez, G. et al. Sparse to dense ground truth preprocessing in hyperspectral imaging for in vivo brain tumour detection. In 2023 IEEE International Conference on Metrology for eXtended Reality, Artificial Intelligence and Neural Engineering (MetroXRAINE), 272–277 (IEEE, 2023).
Martín-Pérez, A. et al. Unifying heterogeneous hyperspectral databases for in vivo human brain cancer classification: Towards robust algorithm development. Computer Methods and Programs in Biomedicine Update 7, 100183, https://doi.org/10.1016/j.cmpbup.2025.100183 (2025).
Kruse, F. A. et al. The spectral image processing system (sips)-interactive visualization and analysis of imaging spectrometer data. Remote sensing of environment 44, 145–163 (1993).
Martín-Pérez, A. et al. SLIMBRAIN Database: A Multimodal Image Database of in vivo Human Brains for Tumor Detection., https://doi.org/10.21950/LAUR3D (2024).
Villa, M. et al. Stitching technique based on surf for hyperspectral pushbroom linescan cameras. In 2021 XXXVI Conference on Design of Circuits and Integrated Systems (DCIS), 1–6, https://doi.org/10.1109/DCIS53048.2021.9666155 (2021).
Allen, M. et al. Raincloud plots: a multi-platform tool for robust data visualization [version 2; peer review: 2 approved]. Wellcome Open Research4, https://doi.org/10.12688/wellcomeopenres.15191.2 (2021).
Fachada, S., Bonatto, D., Schenkel, A. & Lafruit, G. Free navigation in natural scenery with dibr: Rvs and vsrs in mpeg-i standardization. In 2018 International Conference on 3D Immersion (IC3D), 1–6 (IEEE, 2018).
Fachada, S., Bonatto, D., Teratani, M. & Lafruit, G. View synthesis tool for vr immersive video (IntechOpen, 2022).
Chetverikov, D., Svirko, D., Stepanov, D. & Krsek, P. The trimmed iterative closest point algorithm. In 2002 International Conference on Pattern Recognition, vol. 3, 545–548 (IEEE, 2002).
Dziembowski, A., Mieloch, D., Stankowski, J. & Grzelka, A. Iv-psnr–the objective quality metric for immersive video applications. IEEE Transactions on Circuits and Systems for Video Technology (2022).
Acknowledgements
The authors would like to thank the neurosurgeons and staff of the Hospital Universitario 12 de Octubre. This work was supported by the NEMESIS-3D-CM (Intraoperative Classification Of Brain Tumours Through 3D Immersive Models In The Madrid Community) (Y2018/BIO-4826) research project, funded by the Regional Government of Madrid (Spain), by the TALENT-HIPSTER (HIgh Performance Systems and Technologies for E-health and fish faRming) (PID2020-116417RB-C41) research project, funded by the Spanish Ministry of Science and Innovation, and by the European project STRATUM (3D Decision Support Tool for Brain Tumor Surgery) under grant agreement no. 101137416. Additionally, we would like to thank Pedro Lobo for his help in making the SLIMBRAIN database accessible.
Author information
Authors and Affiliations
Contributions
A.M.P. drafted the manuscript; elaborated all the plots, figures and tables; wrote the computer software to acquire, manage, process and upload the hyperspectral data; conceived, analysed and conducted the data validation experiments; assisted the neurosurgeons during the ground-truth labelling procedure; collected the data at the hospital; helped acquire the hyperspectral calibration libraries; designed and built the SLIMBRAIN database website; and recorded the videos for the Usage Notes section; M.V. designed and developed all the acquisition system prototypes; wrote the computer software to acquire and process the hyperspectral, RGB and depth data; conceived the depth image validation experiments; helped calibrate the SLIMBRAIN prototypes 3 and 4; collected the data at the hospital; conceived and helped acquire the hyperspectral calibration libraries; and reviewed the manuscript; G.R.O. designed and developed all the acquisition system prototypes; wrote the computer software to acquire the hyperspectral, RGB and depth data; collected data at the hospital; and helped acquiring the hyperspectral calibration libraries; J.S. wrote computer software to acquire and process RGB and depth data; conceived, conducted and wrote the depth image validation experiments; helped calibrate the SLIMBRAIN prototypes 3 and 4; and helped acquire the hyperspectral calibration libraries; G.V. and G.U. assisted in writing computer software to process the hyperspectral data; helped during data collection at the hospital; and assisted neurosurgeons during the ground-truth labelling procedure; A.M.T. helped build the system to access the database and reviewed the manuscript. M.C. designed and developed all acquisition system prototypes; supervised the research; and reviewed the manuscript; L.J.R., A.P.N. and A.L. helped during the data collection and performed the ground-truth labelling procedure; A.L. reviewed the manuscript and supervised the research; E.J. and C.S. helped in the design and development of all the acquisition system prototypes; supervised the research; and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Martín-Pérez, A., Villa, M., Rosa Olmeda, G. et al. SLIMBRAIN database: A multimodal image database of in vivo human brains for tumour detection. Sci Data 12, 836 (2025). https://doi.org/10.1038/s41597-025-04993-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04993-y