Abstract
Cryo-electron microscopy (cryo-EM) is a transformative imaging technology that enables near-atomic resolution 3D reconstruction of target biomolecule, playing a critical role in structural biology and drug discovery. Cryo-EM faces significant challenges due to its extremely low signal-to-noise ratio (SNR) where the complexity of data processing becomes particularly pronounced. To address this challenge, foundation models have shown great potential in other biological imaging domains. However, their application in cryo-EM has been limited by the lack of large-scale, high-quality datasets. To fill this gap, we introduce CryoCRAB, the first large-scale dataset for cryo-EM foundation models. CryoCRAB includes 746 proteins, comprising 152,385 sets of raw movie frames (116.8 TB in total). To tackle the high-noise nature of cryo-EM data, each movie is split into odd and even frames to generate paired micrographs for denoising tasks. The dataset is stored in HDF5 chunked format, significantly improving random sampling efficiency and training speed. CryoCRAB offers diverse data support for cryo-EM foundation models, enabling advancements in image denoising and general-purpose feature extraction for downstream tasks.
Similar content being viewed by others
Background & Summary
Background
Cryo-electron microscopy (cryo-EM) is a revolutionary biological imaging technology that enables near-atomic resolution 3D structure determination of biomolecules in their native states1,2. This technique has become indispensable in structural biology and drug discovery, particularly for studying macromolecular complexes, protein folding, and viral structures3,4. By rapidly freezing samples, cryo-EM preserves the native conformations of biomolecules, providing a powerful tool for investigating heterogeneous or difficult-to-crystallize targets5,6,7,8.
Cryo-EM Data Processing Workflow
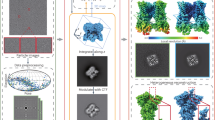
The cryo-EM image processing pipeline consists of a series of critical steps aimed at reconstructing high-resolution 3D structure from raw data. The workflow typically includes motion correction, Contrast Transfer Function (CTF) estimation, micrograph curation, particle picking, and 3D reconstruction (Fig. 1).
Overview of cryo-EM data processing pipeline. The pipeline covers several steps from movie capturing, motion correction, CTF estimation, micrograph curation, particle picking and 3D reconstruction. (a) demonstrates rigid motion, bending motion and patch-wise motion estimation during of movies. (b) shows the 2D fitting data obtained during CTF estimation of micrographs, with the upper part of the image representing reciprocal space and the lower part showing simulated Thon rings. (c) evaluates the quality of images to determine their suitability for reconstruction. (d) accurately picks all particles from the micrograph. (f) performs 2D classification and averaging of the picked particles, removing unsuitable ones. (g) estimates particle poses and performs reconstruction without prior pose information. Finally, (h) refines the reconstructed volume data to achieve high-resolution results.
Motion correction
During cryo-EM imaging, samples are rapidly frozen and then imaged using an electron beam, producing a time-series of images known as movies. Due to noise and sample drift, these movies require motion correction. Motion correction involves aligning and averaging frames from cryo-EM movies to generate a single micrograph, significantly improving SNR by reducing motion blur. During imaging, samples often undergo complex 3D deformations due to electron beam exposure, leading to anisotropic motion9, where different regions of the image shift in varying directions. Traditional methods10,11,12,13,14 such as MotionCor214, rely on frame alignment and image processing algorithms like optical flow and subpixel alignment.
CTF estimation
Contrast transfer function (CTF) estimation involves determining the defocus parameters of the objective lens to correct phase and amplitude modulations during cryo-EM imaging process. The CTF describes how microscope contrast varies with spatial frequency, and its phase inversion effects significantly impact image quality. Uncorrected micrographs can lead to particle phase cancellation, limiting reconstruction resolution. Current methods15,16,17,18,19, such as goCTF17, patch-based CTF18 and CTFFIND416, rely on Thon ring features to match the micrograph’s power spectrum with a CTF model. While they can estimate accurate CTF parameters, there is no existing CTF correction method robust to more comprehensive experimental settings including solution conditions and hardware.
Micrograph curation
Micrograph curation is crucial for high-resolution 3D reconstruction. Due to challenges such as uneven ice thickness, sample impurities, and low SNR, many micrographs are of insufficient quality for high-precision reconstruction. Traditional curation methods rely on CTF estimation results, using thresholds for defocus, astigmatism, and CTF fitting parameters to filter high-quality data.
Particle picking
Particle picking is a critical step in cryo-EM image processing, aiming to locate particles in micrographs for 3D reconstruction. Its accuracy directly impacts reconstruction resolution and efficiency. However, low SNR and sample heterogeneity make this task challenging. Traditional methods, such as DoG Picker20, Xmipp21, AutoCryoPicker22, and KLT Picker23, rely on template matching or local contrast enhancement but struggle with noise and complex samples.
3D reconstruction
3D reconstruction is the final step in cryo-EM processing, where particles are classified and aligned, and a 3D model is constructed. This step requires accurate particle picking and micrograph curation to ensure high-quality reconstructions. Current software tools like CryoSPARC24, RELION14, and Scipion13 offer sophisticated algorithms for 3D reconstruction, with each providing unique strengths in handling noise, particle heterogeneity, and resolution enhancement.
Cryo-EM in the Deep Learning Era
Deep learning has revolutionized scientific imaging, driving breakthroughs that were once considered unattainable. In cryo-EM, it has become a cornerstone for data analysis, enabling significant advancements across the entire image processing pipeline.
Motion correction
Has traditionally been hindered by the extremely low SNR of cryo-EM images, making precise, pixel-level motion estimation a formidable challenge. Recent deep learning-based approaches, such as Noiseflow25 and DST-net26, address these limitations by leveraging synthetic cryo-EM movie data for training. These methods achieve state-of-the-art performance, improving motion correction accuracy and enhancing image quality.
Micrograph curation
Has similarly benefited from deep learning’s ability to improve efficiency, consistency, and accuracy. Automated curation tools, including MicrographCleaner27 and Miffi28, have demonstrated their potential in overcoming challenges like subjective bias and manual inefficiency. For instance, Miffi achieves a 93% higher accuracy than traditional CTF-based methods, setting a new standard for automated micrograph curation.
Particle picking
Remains one of the most critical and challenging steps in cryo-EM processing due to low SNR and sample heterogeneity. Deep learning methods29,30,31,32,33,34,35,36,37,38,39,40, such as APPLE Picker31, crYOLO32, and Topaz33, have significantly enhanced particle localization efficiency and accuracy. Furthermore, recent Transformer-based models like CryoTransformer41, CryoMAE42, and CryoSegNet43 demonstrate remarkable improvements in handling complex samples, setting a new benchmark in particle picking.
Challenges in Constructing Cryo-EM Foundation Model Training Data
Over the past decades, data-driven deep learning methods have achieved unprecedented progress in scientific imaging44,45. Recent advances in foundation models46 have demonstrated remarkable generalization capabilities by leveraging self-supervised learning on large-scale, high-quality datasets47,48. These models have been widely applied to various medical imaging modalities, including CT49,50, X-ray51, ultrasound52, and digital pathology53,54,55,56,57,58,59,60,61, significantly improving downstream task performance.
However, these advancements rely on large-scale, high-quality datasets specific to each imaging modality. Merlin62, a 3D vision language models is trained on a high-quality clinical dataset of paired CT scans (6+ million images from 15,331 CTs), EHR diagnosis codes (1.8+ million codes), and radiology reports (6+ million tokens). UNI61, a general-purpose self-supervised model for pathology, is pretrained using more than 100 million images from over 100,000 diagnostic H&E-stained WSIs (>77 TB of data). RETFound59, a foundation model for retinal images is trained on 1.6 million unlabelled retinal images by means of self-supervised learning and then adapted to disease detection tasks with explicit labels. In cryo-EM, the extremely low signal-to-noise ratio (SNR) and complex sample characteristics11,63,64,65,66,67,68 make it challenging to obtain high-quality images without extensive and meticulous processing. This difficulty has hindered the development of foundation models for cryo-EM.
The largest public repository for cryo-EM data, EMPIAR69, has become a critical resource for algorithm development in the field (see Table 1). The database hosts more than 1,700 single-particle cryo-EM datasets and 10 million cryo-EM movies or micrographs, encompassing diverse stages from sample preparation to image processing. Additionally, the database continues to grow steadily.However, the preprocessed data in EMPIAR comes from diverse sources with inconsistent quality and formats, limiting the efficient construction of high-quality datasets. To address this, we introduce CryoCRAB, the first large-scale, standardized dataset designed for training cryo-EM foundation models. CryoCRAB comprises 152,385 sets of raw movie frames, covering 746 distinct proteins, with a total data volume of 116.8 TB. To generate high-quality micrographs from raw data, we developed an automated processing pipeline incorporating motion correction and Contrast Transfer Function (CTF) estimation. Specifically, to support denoising-related pretraining tasks70, we split each movie into odd and even frames, processing them separately to generate paired micrographs.
Additionally, CryoCRAB offers unique visualization and filtering capabilities. We provide detailed statistical analyses of each micrograph, including intensity distributions, CTF parameters (e.g., resolution and defocus), and pixel motion characteristics. These features enable users to filter subsets of data tailored to specific needs, enhancing training efficiency. To optimize storage and access, CryoCRAB employs the HDF5 format with chunked storage, which significantly improves random sampling efficiency and reduces I/O bottlenecks compared to traditional image formats. This efficient data organization ensures reliable support for large-scale foundation model training and complex computational tasks.
We believe that CryoCRAB will provide diverse data support for foundation model development in cryo-EM, driving the widespread adoption of deep learning in cryo-EM data processing and offering new tools for exploring protein structures and dynamics.
Methods
CryoCRAB is a large-scale, high-quality dataset constructed through a systematic data processing pipeline, specifically designed for training cryo-EM foundation models (see Fig. 2). The pipeline encompasses multiple steps, including data collection, curation, processing, model-training preprocessing, and efficient storing, ensuring both the diversity and quality of the dataset.
Overview of CryoCRAB Dataset. The crucial processing steps includes EMPIAR crawling, motion correction, CTF estimation, micrograph curation, and pre-processing. (a) We crawl file path information and experimental metadata from the EMPIAR database and download the curated movies and gain files. (b) We perform gain correction and motion correction for movies to obtain two types of motion annotations, full-diff micrograph pairs and background estimates. (c) We perform CTF estimation for micrographs to estimate CTF parameters such as defocus value, astigmatism, and phase shift. (d) We curate the processed images based on median intensity, rigid motion statistics, and CTF estimation statistics, which classify the quality of images from 0 to 7. (e) We propose a cryo-EM micrograph pre-processing pipeline to transform the images into the input format required for pre-training models by background subtraction, band-limit CTF filtering, contrast normalization and Z-score standardization.
First, our cryo-EM foundation model dataset is sourced from the EMPIAR public database. The foundation models trained on CryoCRAB are applicable to cryo-EM downstream tasks such as motion correction, CTF estimation, and particle picking, all of which take movies or micrographs as input (see Section 2). We obtained raw cryo-EM movies and associated metadata from EMPIAR, followed by rigorous data cleaning and filtering to ensure reliability and accuracy (see Section 2.1). Subsequently, we processed the data using CryoSPARC with standard steps including motion correction, CTF estimation, and micrograph curation. Notably, we splited each raw movie into odd and even frames, generating full-even-odd micrograph triplets (see Section 2.2).
During preprocessing, we uniformly removed background from images, band-limited the frequency domain to 3Å, applied CTF filtering, and performed contrast normalization to filter low image quality outliers. We also computed the mean and standard deviation of the images for Z-score normalization during training (see Section 2.3). Furthermore, we adopted an efficient storing strategy by converting each full-even-odd micrograph triplet into a full-diff micrograph pair and storing the outlier-removed image data along with normalization parameters in HDF5 format, which significantly improves the data I/O speed, ensuring efficiency during large-scale model training (see Section 2.4).
Input Data Formats for Cryo-EM Foundation Model Pre-training
To construct cryo-EM foundation models, CryoCRAB leverages raw cryo-EM movies as the primary source of image information, addressing the challenge of limited annotations in the field. From these movies, we derive full-even-odd micrograph triplets and generate high-quality annotations, forming the core of the CryoCRAB dataset. This comprehensive approach enables effective pre-training of foundation models, which can be fine-tuned for downstream tasks such as motion correction, CTF estimation, micrograph denoising and micrograph curation.
In cryo-EM workflow, the primary forms of image data are movies, micrographs, and particles. Movies are dynamic image sequences composed of multiple frames, typically captured by direct detector devices (DDDs)71, which offer significantly higher detective quantum efficiency (DQE) compared to traditional cameras. This technology allows cryo-EM to record micrographs as multi-frame movies rather than single-exposure images. These movies capture dynamic information, aiding subsequent processing steps, such as motion correction. Micrographs are generated by corrected movie frames and serve as the basic unit for further analysis. Particles, extracted from micrographs through particle picking techniques, represent individual molecules or fragments used for high-resolution structure reconstruction and modeling. In particular, since the acquisition of particles requires extensive annotation, CryoCRAB does not include particle data in its unannotated dataset.
To enhance the effectiveness of foundation model training, the CryoCRAB dataset not only includes traditional micrographs generated by full-frame averaging but also incorporates even-odd micrographs created by averaging odd and even frames separately. These three types-full, even, and odd micrographs-are collectively referred to as full-even-odd micrograph triplets. This approach is motivated by insights into noisy image restoration, particularly inspired by the Noise2Noise (N2N)72 method, which demonstrates that image denoising can be achieved using only pairs of noisy images. In the context of cryo-EM, these pairs correspond to the even-odd frames derived from the same movie, a concept first effectively utilized for denoising by Topaz73. Leveraging the characteristics of cryo-EM transmission imaging, each full-even-odd micrograph triplet can be efficiently represented as a full micrograph and a difference of even and odd micrographs (diff micrograph), forming a full-diff micrograph pair.
EMPIAR Datasets Collection and Curation
When constructing the foundation model dataset, we sourced cryo-EM data from public databases rather than direct experimental acquisition. EMPIAR is one of the largest cryo-EM data repositories, and we accessed EMPIAR through its REST API to automatically retrieve all EMPIAR entries as well as the associated EMDB74 entries and PDB75 entries. The EMPIAR entries cover basic image information (e.g., image size and pixel size), while the EMDB and PDB entries provide further experimental and structural details (see Fig. 2).
We select 200 raw movies per dataset in order to make a trade-off between storage and diversity. These data are further processed for model training. To meet the requirements for data diversity and high quality in model training, we performed a preliminary filtering process. First, we ensured that the selected datasets are generated using single-particle analysis (SPA) and contain raw movie data, resulting in 746 EMPIAR datasets. On one hand, we aimed to include as many sample preparation and imaging conditions as possible. On the other hand, we recognized the unique value of images captured under different conditions but belonging to the same protein for foundation model training. Therefore, we did not filter datasets by protein type at this stage.
EMPIAR datasets are available in various formats, primarily TIFF, MRC, and EER. TIFF is commonly used for storing multi-frame images with compression, suitable for smaller datasets. MRC stores raw data directly, resulting in larger files that are less storage efficient. EER, supporting thousands of frames, is one of the most primitive formats in cryo-EM. The gain file, which records pixel-specific correction factors, is an essential component of the dataset. However, the formats for gain files in EMPIAR are inconsistent, including DM4, DAT, MRC, and TIFF. Processed gain data is typically stored in MRC or TIFF formats, while raw gain data may be provided in DM4 or DAT formats. To address these inconsistencies and standardize gain data, we utilize the open-source software EMAN2 to convert gain files from DM4 and DAT formats into the MRC format.
These data curation and preprocessing steps ensure that each dataset meets high-quality standards, providing both diversity and consistency for model training.
Processing Movies to Micrographs using CryoSPARC
We employ CryoSPARC for a standardized data processing pipeline. First, we used CryoSPARC’s Patch Motion Correction to align and average movie frames, generating motion-corrected full-even-odd micrograph triplets. Next, we performed CTF estimation to determine key parameters such as defocus, tilt angle, and astigmatism, which are essential for subsequent CTF filtering. Finally, after motion correction and CTF estimation, we curated and annotated the micrographs to ensure high-quality data for foundation model training and downstream task validation.
Motion correction
Motion correction is a critical step in cryo-EM data processing, aimed at compensating for sample displacement caused by radiation pressure, mechanical vibrations, or environmental instability during electron microscopy exposure. This step improves the signal-to-noise ratio (SNR) and contrast of the final images. The motion correction pipeline, as illustrated in Fig. 3, includes: (1) applying gain correction on each frame, (2) estimating motion trajectories from movies, and (3) separately correcting and averaging odd and even frames. CryoCRAB utilizes CryoSPARC’s Patch Motion Correction for this purpose. By integrating experimental parameters from the empiar entries, we generated full-even-odd micrograph triplets from movies and gain files, along with motion data. Notably, leveraging the characteristics of cryo-EM transmission imaging, we converted the full-even-odd micrograph triplet into a full-diff micrograph pair for efficient storage.
Details of Motion Correction Pipeline. (a) The pipeline starts with the input of raw movies and their corresponding gain reference. First, gain correction is applied to address the detector’s non-uniform response. Then, We use the patch motion correction algorithm in CryoSPARC to estimate motion for each frame, followed by motion correction and alignment of even and odd frames separately. Notably, CryoSPARC performs background estimation and background subtraction after the motion correction. Finally, to reduce data storage overhead, the even micrograph is subtracted from the odd micrograph to generate the full-diff MRC pair. (b) The left image shows the patch-wise motion estimation for all frames, with the starting frame in blue and the ending frame in yellow. The global rigid motion and local patch-level bending motion are combined and amplified by a factor of 20 for visualization. The right image displays the pixel-wise optical flow estimation obtained through spline interpolation, as well as the patch-wise motion direction of the current frame relative to the previous frame. (c) We generate the full-even-odd triplet by combining motion-corrected even and odd frames. This approach supports Noise2Noise training. (d) We perform background estimation on the motion-corrected images to separate and subtract the background, enhancing the image contrast.
Gain correction compensates for the non-uniform response of the detector, ensuring uniform and accurate image intensity. The gain file, records correction factors for each pixel. Depending on the detector type, these factors are applied differently. For example, Gatan k2/k3 cameras require dividing raw pixel values by the correction factor, while Falcon 4 cameras require multiplication. Due to the absence of gain reference metadata in EMPIAR, we manually determine the optimal parameters by evaluating all possible flip and rotation combinations for their impact on the uniformity of contrast in the gain-corrected images.
We categorize motion into two types based on its source: (1) Rigid Motion: This refers to mechanical drift of the sample-stage during long-exposure imaging76. Experimental evidence suggests that most observed motion arises from beam-induced bending rather than mechanical instability77. (2) Bending Motion: This is associated with the interaction between the support foil and grid bars due to differential cooling rates during plunge freezing9. The slower cooling of grid bars creates transient tensile stress, which, upon release, can subject the sample to compressive stress, leading to radiation-induced creep and sample warping under electron beam exposure.
For rigid motion, we model the overall sample movement caused by thermal expansion or microscope drift without accounting for anisotropic motion from ice layer changes. This is achieved by iteratively optimizing the full-frame trajectory to maximize inter-frame correlation. For bending motion, we divide the movie into overlapping patches and estimate the displacement of each patch per frame under a spatially and temporally smooth motion model to account for anisotropic motion caused by ice layer changes.
During frame averaging, motion correction results are applied separately to odd and even frames, producing odd-micrographs and even-micrographs. Averaging these yields the full-micrograph. These corrected micrographs effectively eliminate motion blur caused by ice layer changes or equipment vibrations, enhancing contrast and high-frequency information. CryoCRAB records the full-even-odd micrograph triplet, along with the estimated rigid and bending motion data.
CTF estimation
Contrast transfer function (CTF) estimation involves determining the defocus parameters of the objective lens, particularly the defocus, from cryo-EM micrographs. The CTF describes how lens aberrations, including defocus, affect the contrast of the recorded images. By fitting the microscope’s CTF model to the image’s amplitude spectrum, defocus parameters can be estimated, enabling subsequent image correction and processing. This correction is necessary because the CTF introduces frequency-dependent amplitude modulation in the image78.
In single-particle analysis, samples are typically thin (20–100 nm) and can be treated as weak-phase objects77. However, biological macromolecules in solution are primarily composed of light elements, which have a weak effect on the phase of electron waves. As a result, images captured without defocus exhibit minimal or no contrast. Therefore, cryo-EM introduces a controlled amount of defocus during imaging to generate phase contrast. However, due to stage tilt, uneven sample surfaces, or non-uniform particle distribution along the optical axis, the CTF may vary across the micrograph.
CryoCRAB employs CryoSPARC’s Patch CTF Estimation for CTF estimation. As illustrated in Fig. 4, we estimated spatially and temporally smooth defocus distributions for tilted, bent, and deformed samples. The CTF parameter model used by CryoSPARC is consistent with the simplified version in CTFFIND416 for computational efficiency79,80. In subsequent preprocessing steps, we also used this CTF parameter model for CTF filtering to enhance image contrast.
Details of CTF Estimation. (a) The CTF estimation pipeline begins with the input of a micrograph. We first perform initial CTF estimation, followed by calculating the envelope function to correct for attenuation effects. Next, we optimize CTF parameters through 2D CTF estimation and patch-wise CTF refinement, ultimately generating a 2D defocus landscape to visualize the defocus variation across the entire micrograph. (b) We display the 1D search plot, where the peak reflects the optimal defocus value, indicating the best match between the ideal CTF curve and the experimental data at that defocus value. (c) The 1D CTF fit is used to evaluate data quality and the accuracy of CTF fitting. The gray line represents the radial average of the image power spectrum, with its oscillations (Thon rings) reflecting CTF characteristics. The red line shows the ideal CTF curve obtained through patch CTF estimation, representing the average defocus of the micrograph. The blue line indicates the correlation between the power spectrum and the ideal CTF, with the green vertical line marking the CTF fit resolution as a reference for data quality. (d) We analyze the uniformity of the ice layer by estimating the ice thickness. The ice thickness is calculated by comparing the background signal centered at 0.265 Å−1 with a broader frequency band, where a higher background signal typically indicates thicker ice.
The CTF parameter model used by CryoSPARC is a 2D cosine function of the frequency-dependent phase shift χ. g is the spatial frequency vector, and λ is the electron wavelength. Δf1 and Δf2 are the most critical parameters in CTF estimation, used to calculate the microscope’s defocus and astigmatism. The remaining four parameters are optical: Cs is the spherical aberration, Δφ is the additional phase shift introduced by a phase plate (if absent, Δφ = 0), ω is the proportion of total contrast due to amplitude contrast (e.g., electrons scattered outside the objective aperture or energy-filtered81), and α is the azimuthal angle or astigmatism angle, representing the angle between the image x-axis and the Δf1 direction.
Here, ⟨⋅,⋅⟩ denotes the vector inner product, and gx, gy are the x- and y-axis components of the 2D frequency vector g. \(\,{\rm{DF}}\,:\,=\frac{1}{2}(\Delta {f}_{1}+\Delta {f}_{2})\) is the defocus along the optical axis, \(\,{\rm{df}}\,:\,=\frac{1}{2}(\Delta {f}_{1}-\Delta {f}_{2})\) is half the astigmatism along the optical axis, and \({{\rm{df}}}_{xx}\,:\,=\cos (2\alpha ){\rm{df}}\) and \({{\rm{df}}}_{xy}\,:\,=\sin (2\alpha ){\rm{df}}\) represent astigmatism along the Δf1 and Δf2 directions, respectively. We provide a more efficient method for calculating the phase shift χ. Specifically, for the same dataset, we no longer need to recompute defocus for all frequency components when Δf1 and Δf2 change. Instead, we only calculate four variables: DF, df, dfxx, and dfxy, with the latter two depending solely on the azimuthal angle α and not on the frequency component direction.
Cryo-EM samples are often not perfectly “flat”82. Before freezing, particles tend to concentrate near the air-water interface, and the ice surface itself is often irregular83. Since defocus affects the CTF, particles in the same image may have different defocus values, leading to varying contrast transfer functions. We computed a Bézier-curve-smoothed defocus surface by examining multiple regions of the micrograph. First, we performed a coarse CTF estimation assuming no astigmatism on entire micrograph, finding the best-fit defocus by correlating with the radially averaged power spectrum. Next, we used this coarse defocus estimate to compute a new envelope function84, followed by estimating the 2D CTF for the entire micrograph, including astigmatism. Finally, we refined the defocus estimation for each patch, fitting these patch CTF estimates to a spline function to estimate local defocus across the micrograph.
By utilizing non-dose-weighted full micrographs, we derived smooth defocus estimates for specified 2D coordinates, accompanied by a comprehensive set of CTF-related parameters. These parameters include the defocus values Δf1 and Δf2 along the two principal axes, defocus DF along the optical axis, astigmatism df, azimuthal angle α, amplitude contrast proportion ω, and relative ice thickness.
Micrograph curation
The quality of cryo-EM images is influenced by various factors, including sample solution conditions, microscope acquisition parameters, and the target protein type. Existing approaches for cryo-EM image quality assessment, such as the manual labeling scheme proposed by Miffi28, suffer from limitations including incomplete standards, subjective inconsistencies, and a lack of automation. To address these challenges, we designed a comprehensive quality curation scheme based on seven key metrics to annotate each micrograph in a unified and automated manner. These parameters include the Median Intensity of motion-corrected micrographs, the Total Rigid Motion and Total Rigid Motion Curvature derived from motion correction, and four CTF-related statistics: CTF Fit Resolution, Tilt Angle, Defocus Range, and Astigmatism. These metrics help us annotate quality of micrographs, ensuring that high-quality images are selected for subsequent model training. For each metric, we calculate the mean (μ) and standard deviation (σ) across the dataset, using the 3σ interval to determine whether a metric falls within the acceptable range. The image quality score is computed as follows: each metric within the dataset’s acceptable range contributes 1 to the quality score, resulting in a maximum quality score of 7 and a minimum of 0. As shown in Fig. 5, micrographs are categorized into low quality (0–2), medium quality (3–5), and high quality (6–7) based on this scoring system.
Results of Micrograph Curation. We display micrographs categorized into low quality (0–2), medium quality (3–5), and high quality (6–7) based on our quality screening criteria. To evaluate cryo-EM image quality, we design a screening scheme based on seven key parameters, including median intensity after motion correction, total rigid motion and its rate of change (total rigid motion curvature), as well as CTF fit resolution, tilt angle, defocus range, and astigmatism derived from CTF estimation. For each parameter, we calculate the mean (μ) and standard deviation (σ) of the dataset and use the 3σ interval to annotate each parameter. Each parameter within the screening interval contributes 1 point to the image quality score, resulting in a final quality score ranging from 0 to 7. Through this screening process, we have observed a significant improvement in micrograph quality as the quality score increases.
Algorithm 1
Contrast Normalization Algorithm.
Median intensity
The median pixel intensity of each micrograph is calculated to assess overall brightness and contrast. Cryo-EM images often exhibit strong background noise, particularly when the ice layer is uneven, leading to significant variations in noise intensity across regions. By focusing on the median value, this metric effectively mitigates the influence of background noise, ensuring that the evaluation is more representative of protein particle regions. We also observed that protein particle signals in micrographs are typically weaker than background signals. To more accurately estimate protein pixel intensity using the median, we first applied contrast normalization (see Algorithm 1) before computing the median intensity.
Total rigid motion
During motion correction, multiple consecutive frames are aligned and averaged to produce a single micrograph, significantly improving the signal-to-noise ratio. Rigid motion estimation measures the positional offset of each frame relative to a certain frame to achieve maximum global correlation. Total Rigid Motion represents the cumulative rigid motion between adjacent frames in a movie, reflecting the overall displacement of the sample during acquisition. By calculating rigid motion between frames, we can assess sample stability during imaging and identify potential displacement errors or sample drift issues.
Total rigid motion curvature. In addition to Total Rigid Motion, we compute Total Rigid Motion Curvature, which represents the cumulative rate of change in rigid motion between adjacent frames. A high curvature value often indicates significant motion variations during exposure, which tends to degrade image quality. This metric helps identify abrupt or rapidly changing motions that could destabilize the image, providing valuable information for image curation.
CTF fit resolution. We calculate the correlation between the micrograph’s power spectrum and the ideal CTF derived from Patch CTF Estimation. The spatial frequency corresponding to a correlation threshold is defined as the CTF Fit Resolution. This metric is not a hard constraint on the quality of the data but rather than a reference. For example, high-resolution fits may indicate the presence of carbon layers, while low-resolution fits could result from crystalline ice, motion correction failure, or severe radiation damage.
Tilt angle. Unlike “beam tilt” or “coma aberration” in cryo-EM, the Tilt Angle refers to the angle between the tilt axis and the image coordinate axes. To mitigate preferential orientation issues, some SPA experiments collect data with a specific tilt angle applied to the sample stage. However, this angle is not directly input into CryoSPARC during processing but is indirectly estimated through the defocus landscape fitted during Patch CTF estimation. CryoSPARC estimates tilt information by computing the defocus tilt normal vector. Specifically, for a given micrograph, the defocus tilt normal vector [normal[0], normal[1], −1] represents the plane’s normal in 3D coordinates. By normalizing this vector and computing its dot product with the unit normal vector [0, 0, −1], the cosine of the tilt angle is derived, from which the actual tilt angle is calculated.
Defocus range. CryoSPARC’s Patch CTF Estimation measures the defocus landscape across the entire micrograph. The Defocus Range is the difference between the maximum and minimum defocus values within a micrograph, reflecting variations in focus. A large defocus range may indicate uneven focusing, potentially leading to regions with varying contrast in the image.
Astigmatism. Astigmatism, caused by lens asymmetry, typically manifests as focal shifts in certain regions of the image, resulting in elliptical distortion in the frequency domain. Severe astigmatism can blur the image, adversely affecting subsequent reconstruction quality.
Pre-processing of Cryo-EM Micrograph for the Contrast Enhancement
In cryo-EM data processing, preprocessing steps are crucial for enhancing the quality of images used for model training. We employ several key steps during data preprocessing: background subtraction, band-limiting and CTF filtering, contrast normalization, and Z-score standardization. First, we used Gaussian blur to remove background variations caused by uneven ice layers, thereby enhancing the contrast between particles and the background. Next, we applied band-limiting to the frequency domain and performed CTF filtering to reduce the impact of low signal-to-noise ratios and improve image quality. To further enhance visualization, we adjust the pixel value range through contrast normalization, focusing on protein particle regions to improve the contrast between particles and the background. Additionally, we applied Z-score standardization to normalize pixel values to a distribution with zero mean and unit variance, eliminating brightness variations due to acquisition conditions and ensuring data consistency and training stability. These steps lay a solid foundation for subsequent image analysis, particle picking, and model training.
Algorithm 2
Background Subtraction.
Background subtraction
In cryo-EM micrographs, uneven ice thickness affects image processing and the quality of the final reconstructed density maps. Specifically, non-uniform ice distribution leads to variations in contrast between particles and the background across different regions of the micrograph, particularly impacting the accuracy of particle picking models. To address this, we used Gaussian blur to estimate the ice background and subtracted it from the original image, achieving uniform contrast. The implementation details can be found in Algorithm 2. Notably, CryoSPARC’s Patch Motion Correction algorithm automatically performs background subtraction, while other motion correction algorithms without this feature require an additional step for background removal.
Algorithm 3
CTF Filtering.
Band-limit and CTF filtering
To mitigate the impact of low signal-to-noise ratios on model training, we applied band-limiting and CTF filtering to the micrographs. Specifically, we first downsampled the micrographs to a pixel size of about 3 Åto minimize high-frequency noise. Then, using the CTF parameters obtained from CTF estimation, we inverted the CTF before the first peak and applied phase flipping after the first peak. Before the first peak, we multiplied the frequency domain by the reciprocal of the CTF curve; after the first peak, we applied phase flipping by multiplying by the sign of the CTF curve. The implementation details are in Algorithm 3.
Contrast normalization
In cryo-EM data processing, the pixel values in raw cryo-EM movies typically represent the number of electrons detected by the beam at a specific time, usually in integer format. After preprocessing steps such as gain correction and motion correction, the resulting micrograph pixel values represent the corrected cumulative electron dose, typically in floating-point format. However, cryo-EM micrographs often exhibit significant noise and low contrast, making it difficult to distinguish protein particles from the background. To improve visualization, we introduced contrast normalization. The core idea is that protein particle pixel values are concentrated in a narrow range, while background regions (e.g., ice and carbon layers) have extreme pixel values. By adjusting the pixel value range to focus on the protein particle region, we enhance the contrast between particles and the background. The implementation details are in Algorithm 1.
Z-score standardization
In cryo-EM data processing, for each movie image, we independently applied z-score standardization to the full-even-odd triplet obtained from motion correction. The purpose of z-score standardization is to standardize the pixel values of each image to a distribution with zero mean and unit variance, eliminating brightness variations caused by differences in acquisition conditions or background noise. This not only improves numerical stability during training but also accelerates model convergence. Additionally, when using the Noise2Noise loss function for model training, ensuring consistent pixel value distributions across frames or images is critical. If the distributions differ significantly, the model may fail to learn the noise-to-clean mapping effectively, leading to training instability or failure. Through z-score standardization, we ensure data consistency, avoiding training instability due to brightness or noise distribution variations.
The contrast enhancement process from raw movies to preprocessed micrographs is demonstrated in Fig. 6. Each preprocessing step (e.g., background subtraction, band-limiting, CTF correction, contrast normalization) significantly contributes to improving image contrast, further enhancing the quality and effectiveness of the training data. By comparing preprocessed and original images, we can clearly see how each processing step enhances image quality, providing better input data for training foundational models. While contrast improvements can aid in training some deep learning models, contrast normalization or CTF-filtered micrographs may not be ideal for cryo-EM tasks related to particle reconstruction. Contrast normalization, for example, can lead to information loss by clipping image values into a narrower range. Additionally, traditional reconstruction methods, such as RELION14, do not apply CTF correction directly to the images. Instead, they backproject the effect of the CTF to volume space and use the CTF to regularize the reconstructed 3D volume. For these reasons, we recommend that readers avoid using contrast-enhanced micrographs for reconstruction-related cryo-EM tasks.
Validation details of CryoCRAB. The left column shows the micrograph images and their corresponding frequency domain representations, while the right column displays the intensity histograms of the images along with their minimum and maximum values. The preprocessing pipeline includes the following steps: (a) Input raw images: Displays the original unprocessed images and their frequency domain characteristics. (b) Background subtraction: Eliminates background variations caused by ice layer inhomogeneity using Gaussian blur, enhancing the contrast between signals and the background. (c) Band-limit to 3 Å: Applies band-limiting to the frequency domain of the images, reducing the impact of low signal-to-noise ratio regions and improving image quality. (d) CTF Filtered: Applies CTF filtering to further optimize the signal-to-noise ratio of the images. (e) Normalization and Standardization: Adjusts the pixel value range through contrast normalization, focusing on protein particle regions, followed by Z-score standardization to transform pixel values into a distribution with zero mean and unit variance, eliminating brightness bias and ensuring data consistency.
Storing Data into Half-Precision Full-Diff Micrograph Pairs in HDF5
In cryo-EM data storage, efficiently managing large volumes of micrograph data is a critical challenge. To address this, we propose an optimized data storage strategy aimed at reducing storage requirements and improving data access efficiency. First, we converted each full-even-odd triplet into a full-diff micrograph pair, leveraging the relationships “full = even + odd” and “diff = even - odd” to save one-third of the storage space. Next, we used the HDF5 format for data storage, which supports large-scale data management and random access. The chunked storage and compression capabilities of HDF5 significantly accelerate data reading while reducing storage space. Additionally, we converted data from single-precision float format to half-precision float format, maintaining image quality while further reducing storage requirements and I/O overhead. These techniques provide an efficient and scalable storage solution for large-scale data processing and model training.
Half-Precision Block Storage
To accommodate the random cropping strategy commonly used in large-scale model training, we adopted a storage scheme better suited for high-resolution images. Traditional cryo-EM micrographs are typically stored in MRC files following the CCP4 format, but MRC format suffers from significant I/O bottlenecks during large-scale random access, which can severely impact training efficiency. Therefore, we introduced HDF5-based storage, which supports efficient chunked storage and significantly accelerates random access. Additionally, HDF5 supports various compression methods to reduce storage space and is compatible with multiple data types, allowing the embedding of metadata (e.g., annotations and statistics) for easier data management and usage. Furthermore, we optimized data precision. Experiments show that converting micrographs from float32 to half-precision float16 format has minimal impact on 3D reconstruction resolution and image details. Therefore, to further reduce storage requirements, we stored all data in float16 format, ensuring data quality while significantly reducing storage space and I/O overhead.
Data Records
The dataset is available at ScienceDB (https://doi.org/10.57760/sciencedb.17922)85. CryoCRAB comprises 152,385 sets of raw movie frames, covering 746 datasets from EMPIAR. Each EMPIAR dataset typically includes approximately 200 cryo-EM images, consisting of raw movies, motion-corrected full-diff micrographs in MRC format along with estimated background images, and preprocessed full-diff micrographs in HDF5 format (see Fig. 2). The entire dataset, including micrographs and metadata from empiar entries86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258,259,260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318,319,320,321,322,323,324,325,326,327,328,329,330,331,332,333,334,335,336,337,338,339,340,341,342,343,344,345,346,347,348,349,350,351,352,353,354,355,356,357,358,359,360,361,362,363,364,365,366,367,368,369,370,371,372,373,374,375,376,377,378,379,380,381,382,383,384,385,386,387,388,389,390,391,392,393,394,395,396,397,398,399,400,401,402,403,404,405,406,407,408,409,410,411,412,413,414,415,416,417,418,419,420,421,422,423,424,425,426,427,428,429,430,431,432,433,434,435,436,437,438,439,440,441,442,443,444,445,446,447,448,449,450,451,452,453,454,455,456,457,458,459,460,461,462,463,464,465,466,467,468,469,470,471,472,473,474,475,476,477,478,479,480,481,482,483,484,485,486,487,488,489,490,491,492,493,494,495,496,497,498,499,500,501,502,503,504,505,506,507,508,509,510,511,512,513,514,515,516,517,518,519,520,521,522,523,524,525,526,527,528,529,530,531,532,533,534,535,536,537,538,539,540,541,542,543, totals approximately 12.18 TB.
Due to the substantial storage size of raw cryo-EM movie data (86.2 TB), which far exceeds the size of the micrograph portion, we have not uploaded the movie data to conserve storage resources and avoid redundancy. However, the metadata files for the micrographs include FTP paths to the original movies and gain references on EMPIAR, allowing users to download them as needed.
The file directory is organized by EMPIAR ID, with each folder containing subfolders named “micrograph” for MRC-format micrographs, “micrograph_h5” for HDF5-format micrographs, and “background” for estimated background images. Additionally, JSON-format metadata files are provided for each dataset.
Movie
The raw cryo-EM data is stored in EMPIAR, with common file formats including MRC, TIFF, and EER. Corresponding gain reference files are provided for gain correction to eliminate the impact of optical artifacts on image contrast.
Micrograph
Raw movies are processed using CryoSPARC’s Patch Motion Correction module to generate motion-corrected micrographs, with the sample background estimated via Gaussian blur subtracted. To support downstream denoising tasks, CryoCRAB further optimizes the MRC-format data by storing it as Float16-precision full-diff micrographs, achieving data compression while preserving image quality.
These micrographs have undergone CryoSPARC Patch Motion Correction but have not been processed further. They are considered raw micrographs, intended as input for single-particle analysis. Since these micrographs are stored in float16 format, they may not be properly visualized by some commonly used software tools, such as IMOD. To visualize them, we recommend using a custom approach like: (1) read the micrograph using the mrcfile library in Python, and (2) display it using matplotlib with a grayscale colormap.
Micrograph_h5
To enhance micrograph contrast and reduce the impact of high-frequency noise on foundation model training, we applied a series of preprocessing steps to the raw micrographs. These steps include frequency-domain band-limiting, CTF correction, contrast normalization, and Z-score standardization. The final images are stored in HDF5 format as chunked Float16-precision full-diff pairs, significantly improving data loading speed and optimizing the efficiency of foundation model training.
Background
To eliminate the influence of sample background on micrograph contrast, CryoCRAB uses a Gaussian blur-based algorithm to estimate and subtract the background from raw micrographs. We also provide the estimated background images, which can be used for training tasks requiring background-inclusive data or to reconstruct raw micrographs with background through simple operations. Traditional compression methods reduce the background images to a few KB, minimizing storage usage.
Metadata
CryoCRAB records the parameters and results from the processing pipeline for each movie in JSON format. The metadata includes optical parameters used during acquisition, FTP paths to the original movies and gain references on EMPIAR, as well as processed image dimensions, pixel size, storage size, and relative paths within the CryoCRAB dataset. Specifically, the metadata stores rigid motion and bending motion generated during motion correction, along with CTF estimation parameters such as defocus and astigmatism. We recommend importing CryoCRAB metadata into MongoDB and using MongoDB Compass for GUI-based querying and management to enhance usability and efficiency.
3D Reconstruction Metadata
In addition to the image-level metadata generated during image processing, CryoCRAB also provides the corresponding EMDB entry ID for each EMPIAR dataset. This information is stored in the ’cryocrab_emdb.csv’ file alongside the datasets, making it easy to access 3D reconstructions from the public EMDB74 data bank using the provided EMDB ID.
Technical Validation
Analysis of Training DRACO on CryoCRAB
To validate the correctness and the diversity of CryoCRAB data processing pipeline, we present the number of outliers for each metric in Fig. 7(a), the distributions of the seven curation metrics in Fig. 7 (b ~ h), and the distribution of dataset quality scores in Fig. 7(i). To demonstrate that CryoCRAB is of sufficient quality to support the training of cryo-EM foundation models, we trained the cryo-EM foundation model DRACO70 on CryoCRAB and validated its performance on the downstream task of Micrograph Denoising.
Qualitative evaluation of CryoCRAB’s quality on curation statistics. We demonstrate the number of outliers for each curation metric (a) and the distributions of seven curation statistics (b–h), including: Median Intensity (b), Total Rigid Motion (c), Total Rigid Motion Curvature (d), CTF Fit Resolution (e), Tilt Angle (f), Astigmatism (g), and Defocus Range (h). Additionally, we show the distribution of quality scores for the dataset (i).
DRACO, trained on CryoCRAB by dividing full-even-odd micrograph triplets for simultaneous mask autoencoder (MAE)70 pretraining and N2N denoising, demonstrates robust feature extraction capabilities and strong generalization in downstream tasks such as Micrograph Denoising. The training pipeline is shown in Fig. 8. We divided all micrographs with Quality = 7 in CryoCRAB into training and validation sets, with 5 images randomly selected from each dataset to form a validation set of 3,730 images, and the remaining images forming a training set of 139,594 images. Using the same hyperparameters as in the original DRACO paper, we trained DRACO for 200 epochs on two versions of CryoCRAB: Bin1MRC, which includes only background subtraction and is stored in MRC format at the original resolution, and Bin3ÅH5, which undergoes full preprocessing (including frequency-domain truncation to 3Å) and is stored in chunked HDF5 format. The DRACO model trained on Bin1MRC is denoted as DRACO Bin1, and the one trained on Bin3ÅH5 is denoted as DRACO Bin3Å.
Qualitative evaluation of CryoCRAB’s quality on DRACO’s pre-training. (a) We illustrate the training pipeline of DRACO, which includes MAE pre-training and Noise2Noise (N2N) denoising using full-even-odd micrograph triplets, resulting in a robust cryo-EM image feature extractor with strong generalization capabilities. (b) We compare the original images and denoising results of Bin1MRC and Bin3Å data. Bin3ÅH5 data enhances low-frequency information through frequency domain downsampling to 3 Å and bandpass CTF filtering, significantly improving image contrast after denoising, making it more suitable for visual inspection tasks such as particle picking. In contrast, Bin1MRC data retains the original resolution, making it more suitable for reconstruction tasks. (c) We show the training loss curves and training time comparison of Bin1MRC and Bin3ÅH5 data on the DRACO model. The training loss of Bin3ÅH5 is significantly lower than that of Bin1MRC, and the training time is reduced by nearly 8 times, indicating that the preprocessing pipeline significantly accelerates disk I/O and improves training speed.
In Fig. 8(b), we compare the denoising performance of DRACO Bin1 and DRACO Bin3Å. It is evident that DRACO Bin3Å produces micrographs with higher contrast, as the preprocessing steps in CryoCRAB enhance low-frequency information through downsampling to 3Å and correct phase flipping caused by the CTF during imaging. This correction helps the network distinguish between background and protein particle signals more effectively. In summary, Bin3ÅH5 data is more suitable for visual inspection tasks like particle picking, where resolution is not strictly required, while Bin1MRC data, retaining the original resolution, is better suited for reconstruction-related downstream tasks.
In Fig. 8(c), we present the loss curves for DRACO Bin1 and DRACO Bin3Å during training, as well as a comparison of training times for 200 epochs. We observe that DRACO Bin3Å achieves significantly lower training loss, indicating that Bin3Å data contains richer information, enabling the network to converge to a better solution. Additionally, DRACO Bin3Å reduces training time by nearly six times compared to DRACO Bin1, demonstrating that the preprocessing pipeline in CryoCRAB significantly accelerates disk I/O during training, improving overall training efficiency.
Analysis of Even-Odd Micrographs in CryoCRAB
CryoCRAB consists of even-odd micrograph pairs, which are generated by applying consistent motion correction to the even and odd frames of cryo-EM movies. These pairs are particularly useful for cryo-EM tasks that involve noise, such as micrograph denoising73 and training denoising-reconstruction models70. Each even-odd pair contains the same signal (e.g., protein particles, vitreous ice), but the noise differs between the pairs, while remaining consistently distributed. We illustrate this by calculating the consistent signal-to-noise ratio (SNR) for the even-odd micrograph pairs in CryoCRAB from 746 datasets.
Even-odd pair modeling
Consider a framed-averaged cryo-EM micrograph with dimensions m by n. We define its 1-D flattened representation M = S + N as a vector of pixels, where the signal vector \(S \sim {{\mathbb{R}}}^{m\times n}\) and the noise vector \(N \sim {{\mathbb{R}}}^{m\times n}\) represent the underlying signal and additive noise, respectively. The noise N encompasses all types of signal-independent noise, such as detector shot noise. Since the even-odd pair contains the same signal, we represent the even micrograph as Me = S + Ne and the odd micrograph as Mo = S + No, where the only difference is the i.i.d. noise components Ne and No.
SNR of even-odd micrograph pairs
The SNR of a micrograph is defined as the ratio of the signal variance to the noise variance: \(\,{\rm{SNR}}(M)=\frac{{\rm{Var}}(S)}{{\rm{Var}}(N)}\), where Var(S) and Var(N) represent the variances of the signal and noise, respectively. Under the two key assumptions that (1) the signal and noise are independent, and (2) the noise in the even and odd frames is i.i.d., we can derive the following:
Here, Cov[X, Y] represents the covariance between two images and is computed as \(\,{\rm{Cov}}\,[X,Y]=\frac{1}{mn}{\sum }_{i}^{mn}\)\(({X}_{i}-\bar{X})({Y}_{i}-\bar{Y})\). Using this formula, we can calculate the signal-to-noise ratio (SNR) of the even and odd micrographs. For the even micrograph, the SNR is given by \(\,{\rm{SNR}}({M}_{e})=\frac{{\rm{Var}}(S)}{{\rm{Var}}\,({N}_{e})}\), and for the odd micrograph, it is \(\,{\rm{SNR}}({M}_{o})=\frac{{\rm{Var}}(S)}{{\rm{Var}}\,({N}_{o})}\).
Additionally, based on the two assumptions about cryo-EM noise, we can express the SNR of the full micrographs in an equivalent form as \(\,{\rm{SNR}}(M)=\frac{{\rm{Var}}(S)}{{\rm{Var}}(N)}\). This leads to the following equation for the overall SNR of the full micrographs:
To analyze the relationship between even and odd micrographs, we examined 7,430 Bin1MRC full-diff pairs and generated an even-odd SNR scatter plot (Fig. 9(a)). The data revealed a linear relationship described by SNR(Me) ≈ kSNR(Mo) with k = 1.01. From this observation, we draw two conclusions: (1) because of the radiation damage caused by the electron beam, the smaller numbered frames should have a higher signal-to-noise ratio. Even micrographs demonstrate consistently higher SNR values compared to their odd counterparts, indicating a systematic quality difference between these subsets. This consistent bias (approximately 2.4%) likely results from beam-induced specimen damage during the acquisition sequence. (2) despite this quality differential, the strong linear correlation between even and odd micrograph SNRs confirms that both subsets preserve comparable structural information, thus supporting the established practice of utilizing even-odd pairs for effective micrograph denoising.
Quantitative evaluation of CryoCRAB’s SNR distribution. (a) Scatter plot comparing the SNR of even and odd micrographs, demonstrating a consistent linear relationship with similar SNR values for both. (b) Scatter plot for full micrographs vs. even-odd micrographs, illustrating that full micrographs have a higher SNR, approximately twice the value of even-odd micrographs. (c) Histograms of the SNR (in dB) distribution for full, even, and odd micrographs, showing that the SNR of cryo-EM micrographs follows a Gaussian distribution, where SNR in dB is calculated by \({{\rm{SNR}}}_{db}=10{\log }_{10}({\rm{SNR}})\).
SNR relation between full and even-odd micrographs
Raw cryo-EM images are captured using high-speed (~100 FPS) direct detector devices (DDD) as stacks of frames. Full micrographs are generated by averaging all motion-corrected frames, while even-odd micrographs utilize only half of the frames (either even or odd frames). Due to this difference in frame averaging, noise is reduced approximately twice as much in full micrographs compared to even-odd micrographs. To quantify this relationship, we analyzed SNR scatter plots for full-even and full-odd micrograph pairs as shown in Fig. 9(b). The plots demonstrate that full micrographs consistently exhibit an SNR that is approximately twice (one time higher than) that of even-odd micrographs, which aligns with theoretical expectations based on noise reduction properties of frame averaging.
Gaussian estimation of cryo-EM SNR distribution
We present three histograms of the SNR for CryoCRAB full-even-odd micrographs in Fig. 9(c). The histograms show that the SNR of cryo-EM micrographs can be well approximated by the Gaussian distribution, where \(\,{\rm{SNR}}(M) \sim {\mathcal{N}}(\mu ,\sigma )\), \(\,{\rm{SNR}}\,\,({M}_{e}) \sim {\mathcal{N}}({\mu }_{e},{\sigma }_{e})\) and \(\,{\rm{SNR}}({M}_{o}) \sim {\mathcal{N}}({\mu }_{o},{\sigma }_{o})\). This Gaussian behavior is consistent across all three micrograph types, with the full micrographs exhibiting a higher mean SNR value (μ) compared to even (μe) and odd (μo) micrographs. The standard deviations (σ, σe, σo) exhibit minimal variation due to the properties of the logarithmic transformation applied on raw SNR.
Code availability
The scripts used to construct the CryoCRAB dataset are open-sourced at (https://github.com/Dylan8527/CryoCRAB). These include scripts for cleaning, filtering, and downloading EMPIAR datasets, automating CryoSPARC workflows, motion correction for movies, CTF correction for micrographs, and preprocessing for CryoCRAB.
References
Yip, K. M., Fischer, N., Paknia, E., Chari, A. & Stark, H. Atomic-resolution protein structure determination by cryo-em. Nature 587, 157–161 (2020).
Nakane, T. et al. Single-particle cryo-em at atomic resolution. Nature 587, 152–156 (2020).
Huang, X., Luan, B., Wu, J. & Shi, Y. An atomic structure of the human 26s proteasome. Nature structural & molecular biology 23, 778–785 (2016).
Cheng, Y. Single-particle cryo-em-how did it get here and where will it go. Science 361, 876–880 (2018).
Danev, R., Yanagisawa, H. & Kikkawa, M. Cryo-electron microscopy methodology: current aspects and future directions. Trends in biochemical sciences 44, 837–848 (2019).
Cheng, Y., Grigorieff, N., Penczek, P. A. & Walz, T. A primer to single-particle cryo-electron microscopy. Cell 161, 438–449 (2015).
García-Nafría, J. & Tate, C. G. Cryo-electron microscopy: moving beyond x-ray crystal structures for drug receptors and drug development. Annual review of pharmacology and toxicology 60, 51–71 (2020).
Casasanta, M. A. et al. Microchip-based structure determination of low-molecular weight proteins using cryo-electron microscopy. Nanoscale 13, 7285–7293 (2021).
Thorne, R. E. Hypothesis for a mechanism of beam-induced motion in cryo-electron microscopy. IUCrJ 7, 416–421 (2020).
Abrishami, V. et al. Alignment of direct detection device micrographs using a robust optical flow approach. Journal of Structural Biology 189, 163–176 (2015).
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-em using a 2.6 å reconstruction of rotavirus vp6. eLife 4, e06980 (2015).
Rubinstein, J. L. & Brubaker, M. A. Alignment of cryo-em movies of individual particles by optimization of image translations. Journal of Structural Biology 192, 188–195 (2015).
Střelák, D., Filipovič, J., Jiménez-Moreno, A., Carazo, J. & Sánchez Sorzano, C. Flexalign: An accurate and fast algorithm for movie alignment in cryo-electron microscopy. Electronics 9, 1040 (2020).
Zheng, S. Q. et al. Motioncor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature Methods 14, 331–332 (2017).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of Structural Biology 142, 334–347 (2003).
Rohou, A. & Grigorieff, N. Ctffind4: Fast and accurate defocus estimation from electron micrographs. Journal of Structural Biology 192, 216–221 (2015).
Su, M. goctf: Geometrically optimized ctf determination for single-particle cryo-em. Journal of Structural Biology 205, 22–29 (2019).
Tegunov, D. & Cramer, P. Real-time cryo-electron microscopy data preprocessing with warp. Nature Methods 16, 1146–1152 (2019).
Turoňová, B., Schur, F. K., Wan, W. & Briggs, J. A. Efficient 3d-ctf correction for cryo-electron tomography using novactf improves subtomogram averaging resolution to 3.4 å. Journal of Structural Biology 199, 187–195 (2017).
Voss, N., Yoshioka, C., Radermacher, M., Potter, C. & Carragher, B. Dog picker and tiltpicker: Software tools to facilitate particle selection in single particle electron microscopy. Journal of Structural Biology 166, 205–213 (2009).
Abrishami, V. et al. a pattern matching approach to the automatic selection of particles from low-contrast electron micrographs. Bioinformatics 29, 2460–2468 (2013).
Al-Azzawi, A., Ouadou, A., Tanner, J. J. & Cheng, J. Autocryopicker: An unsupervised learning approach for fully automated single particle picking in cryo-em images. BMC Bioinformatics 20, 326 (2019).
Eldar, A., Landa, B. & Shkolnisky, Y. Klt picker: Particle picking using data-driven optimal templates. Journal of Structural Biology 210, 107473 (2020).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryosparc: Algorithms for rapid unsupervised cryo-em structure determination. Nature Methods 14, 290–296 (2017).
Chong, X., Zhou, N., Li, Q. & Leung, H. Noiseflow: Learning optical flow from low snr cryo-em movie. In 2022 26th International Conference on Pattern Recognition (ICPR), 3471–3477 (IEEE, Montreal, QC, Canada, 2022).
Chong, X., Leung, H., Li, Q., Yao, J. & Zhou, N. Deep spatio-temporal network for low snr cryo-em movie frame enhancement. IEEE/ACM Transactions on Computational Biology and Bioinformatics 1–12 (2024).
Sanchez-Garcia, R., Segura, J., Maluenda, D., Sorzano, C. & Carazo, J. Micrographcleaner: A python package for cryo-em micrograph cleaning using deep learning. Journal of Structural Biology 210, 107498 (2020).
Xu, D. & Ando, N. Miffi: Improving the accuracy of cnn-based cryo-em micrograph filtering with fine-tuning and fourier space information. Journal of Structural Biology 216, 108072 (2024).
Wang, F. et al. Deeppicker: A deep learning approach for fully automated particle picking in cryo-em. Journal of Structural Biology 195, 325–336 (2016).
Xiao, Y. & Yang, G. A fast method for particle picking in cryo-electron micrographs based on fast r-cnn. In APPLIED MATHEMATICS AND COMPUTER SCIENCE: Proceedings of the 1st International Conference on Applied Mathematics and Computer Science, 020080 (Rome, Italy, 2017).
Heimowitz, A., Andén, J. & Singer, A. Apple picker: Automatic particle picking, a low-effort cryo-em framework. Journal of Structural Biology 204, 215–227 (2018).
Wagner, T. et al. Sphire-cryolo is a fast and accurate fully automated particle picker for cryo-em. Communications Biology 2, 218 (2019).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nature Methods 16, 1153–1160 (2019).
Zhang, J. et al. Pixer: An automated particle-selection method based on segmentation using a deep neural network. BMC Bioinformatics 20, 41 (2019).
Yao, R., Qian, J. & Huang, Q. Deep-learning with synthetic data enables automated picking of cryo-em particle images of biological macromolecules. Bioinformatics 36, 1252–1259 (2020).
Al-Azzawi, A. et al. Deepcryopicker: Fully automated deep neural network for single protein particle picking in cryo-em. BMC Bioinformatics 21, 509 (2020).
George, B. et al. Cassper is a semantic segmentation-based particle picking algorithm for single-particle cryo-electron microscopy. Communications Biology 4, 200 (2021).
Nguyen, N. P., Ersoy, I., Gotberg, J., Bunyak, F. & White, T. A. Drpnet: Automated particle picking in cryo-electron micrographs using deep regression. BMC Bioinformatics 22, 55 (2021).
Zhang, X., Zhao, T., Chen, J., Shen, Y. & Li, X. Epicker is an exemplar-based continual learning approach for knowledge accumulation in cryoem particle picking. Nature Communications 13, 2468 (2022).
Li, S., Li, H., Zhang, C., Zhang, F. & Wan, X. A segmentation-aware synergy network for single particle recognition in cryo-em. In 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 1066–1071 (IEEE, Las Vegas, NV, USA, 2022).
Dhakal, A., Gyawali, R., Wang, L. & Cheng, J. Cryotransformer: A transformer model for picking protein particles from cryo-em micrographs. Bioinformatics 40, btae109 (2024).
Xu, C., Zhan, X. & Xu, M. Cryomae: Few-shot cryo-em particle picking with masked autoencoders (2024).
Gyawali, R., Dhakal, A., Wang, L. & Cheng, J. Cryosegnet: Accurate cryo-em protein particle picking by integrating the foundational ai image segmentation model and attention-gated u-net. Briefings in Bioinformatics 25, bbae282 (2024).
Anton, J. et al. How well do self-supervised models transfer to medical imaging? Journal of Imaging 8, 320 (2022).
Chen, L. et al. Self-supervised learning for medical image analysis using image context restoration. Medical image analysis 58, 101539 (2019).
Bommasani, R. et al. On the opportunities and risks of foundation models. arXiv preprint arXiv:2108.07258 (2021).
Radford, A. et al. Learning transferable visual models from natural language supervision. In International conference on machine learning, 8748–8763 (PMLR, 2021).
Brown, T. B. et al. Language models are few-shot learners. Adv. Neural Inform. Process. Syst. 33, 1877–1901 (2020).
Hamamci, I. E. et al. A foundation model utilizing chest ct volumes and radiology reports for supervised-level zero-shot detection of abnormalities (2024).
Tölle, M. et al. Federated foundation model for cardiac ct imaging (2024).
Bluethgen, C. et al. A vision–language foundation model for the generation of realistic chest x-ray images. Nature Biomedical Engineering (2024).
Jiao, J. et al. Usfm: A universal ultrasound foundation model generalized to tasks and organs towards label efficient image analysis (2024).
Lu, M. Y. et al. A visual-language foundation model for computational pathology. Nature Medicine 30, 863–874 (2024).
Huang, Z., Bianchi, F., Yuksekgonul, M., Montine, T. J. & Zou, J. A visual–language foundation model for pathology image analysis using medical twitter. Nature Medicine 29, 2307–2316 (2023).
Xu, H. et al. A whole-slide foundation model for digital pathology from real-world data. Nature 630, 181–188 (2024).
Pai, S. et al. Foundation model for cancer imaging biomarkers. Nature Machine Intelligence 6, 354–367 (2024).
Ma, C., Tan, W., He, R. & Yan, B. Pretraining a foundation model for generalizable fluorescence microscopy-based image restoration. Nature Methods 21, 1558–1567 (2024).
Campanella, G., Vanderbilt, C. & Fuchs, T. Computational pathology at health system scale - self-supervised foundation models from billions of images. AAAI 2024 Spring Symposium on Clinical Foundation Models (2024).
Zhou, Y. et al. A foundation model for generalizable disease detection from retinal images. Nature 622, 156–163 (2023).
Wang, X. et al. A pathology foundation model for cancer diagnosis and prognosis prediction. Nature (2024).
Chen, R. J. et al. Towards a general-purpose foundation model for computational pathology. Nature Medicine 30, 850–862 (2024).
Blankemeier, L. et al. Merlin: A vision language foundation model for 3d computed tomography (2024).
Palovcak, E., Asarnow, D., Campbell, M. G., Yu, Z. & Cheng, Y. Enhancing the signal-to-noise ratio and generating contrast for cryo-em images with convolutional neural networks. IUCrJ 7, 1142–1150 (2020).
Elmlund, H., Elmlund, D. & Bengio, S. Prime: probabilistic initial 3d model generation for single-particle cryo-electron microscopy. Structure 21, 1299–1306 (2013).
Li, H. et al. Noise-transfer2clean: denoising cryo-em images based on noise modeling and transfer. Bioinformatics 38, 2022–2029 (2022).
Baxter, W. T., Grassucci, R. A., Gao, H. & Frank, J. Determination of signal-to-noise ratios and spectral snrs in cryo-em low-dose imaging of molecules. Journal of structural biology 166, 126–132 (2009).
Sindelar, C. V. & Grigorieff, N. Optimal noise reduction in 3d reconstructions of single particles using a volume-normalized filter. Journal of structural biology 180, 26–38 (2012).
Scheres, S. H. A bayesian view on cryo-em structure determination. Journal of molecular biology 415, 406–418 (2012).
Iudin, A. et al. Empiar: The electron microscopy public image archive. Nucleic Acids Research 51, D1503–D1511 (2023).
Shen, Y. et al. Draco: A denoising-reconstruction autoencoder for cryo-em. In Globerson, A. et al. (eds.) Advances in Neural Information Processing Systems, vol. 37, 23630–23654, https://proceedings.neurips.cc/paper_files/paper/2024/file/2a1e2162d17c4986934d7740255c0157-Paper-Conference.pdf (Curran Associates, Inc., 2024).
Reimer, L. & Kohl, H. Transmission electron microscopy : physics of image formation. Springer series in optical sciences 36, fifth edition. edn. (Springer, New York, New York, 2008).
Lehtinen, J. et al. Noise2Noise: Learning image restoration without clean data. Proc. Mach. Learn. Res. 80, 2965-2974 (2018).
Bepler, T., Kelley, K., Noble, A. J. & Berger, B. Topaz-denoise: General deep denoising models for cryoem and cryoet. Nature Communications 11, 5208 (2020).
The wwPDB Consortium. et al. Emdb—the electron microscopy data bank. Nucleic Acids Research 52, D456–D465 (2024).
Berman, H. M. et al. The protein data bank. Nucleic Acids Research 28, 235–242 (2000).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-em. Nature methods 10, 584–590 (2013).
Zhang, Y. et al. Single-particle cryo-em: alternative schemes to improve dose efficiency. Journal of Synchrotron Radiation 28, 1343–1356 (2021).
Thon, F. Zur defokussierungsabhängigkeit des phasenkontrastes bei der elektronenmikroskopischen abbildung. Zeitschrift für Naturforschung A 21, 476–478 (1966).
van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. A new generation of the imagic image processing system. Journal of structural biology 116, 17–24 (1996).
Fernando, K. V. & Fuller, S. D. Determination of astigmatism in tem images. Journal of structural biology 157, 189–200 (2007).
Yonekura, K., Braunfeld, M. B., Maki-Yonekura, S. & Agard, D. A. Electron energy filtering significantly improves amplitude contrast of frozen-hydrated protein at 300 kv. Journal of structural biology 156, 524–536 (2006).
Noble, A. J. et al. Routine single particle cryoem sample and grid characterization by tomography. Elife 7, e34257 (2018).
Singer, A. & Sigworth, F. J. Computational methods for single-particle electron cryomicroscopy. Annual review of biomedical data science 3, 163–190 (2020).
Pennycook, S. J. Transmission electron microscopy: A textbook for materials science, williams david b and carter c barry. springer, new york, 2009, 932 pages. isbn 978-0-387-76500-6 (hardcover), isbn 978-0-387-76502-0 (softcover) (2010).
Qihe, C. & Yuan, P. Cryocrab: A large-scale curated and filterable dataset for cryo-em foundation model pre-training. https://doi.org/10.57760/sciencedb.17922 (2024).
Bai, X.-c., Fernandez, I. S., McMullan, G. & Scheres, S. H. Ribosome structures to near-atomic resolution from thirty thousand cryo-em particles. eLife 2, https://doi.org/10.7554/eLife.00461 (2013).
Allegretti, M. et al. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an f-type atp synthase. Nature 521, 237–240, https://doi.org/10.1038/nature14185 (2015).
Danev, R. & Baumeister, W. Cryo-em single particle analysis with the volta phase plate. eLife 5, https://doi.org/10.7554/eLife.13046 (2016).
Danev, R., Tegunov, D. & Baumeister, W. Using the volta phase plate with defocus for cryo-em single particle analysis https://doi.org/10.1101/085530 (2016).
M, K., M, R., W, B. & R, D. Cryo-em structure of haemoglobin at 3.2 Å determined with the volta phase plate https://doi.org/10.6019/EMPIAR-10084 (2016).
Laurinmäki, P. et al. Structure of nora virus at 2.7 Å resolution and implications for receptor binding, capsid stability and taxonomy. Scientific Reports 10, https://doi.org/10.1038/s41598-020-76613-1 (2020).
Schmid, M. et al. Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control. Nature Communications 9, https://doi.org/10.1038/s41467-017-02707-6 (2018).
von Loeffelholz, O. et al. Volta phase plate data collection facilitates image processing and cryo-em structure determination. Journal of Structural Biology 202, 191–199, https://doi.org/10.1016/j.jsb.2018.01.003 (2018).
Herzik, M. A., Wu, M. & Lander, G. C. Achieving better-than-3-Å resolution by single-particle cryo-em at 200 kev. Nature Methods 14, 1075–1078, https://doi.org/10.1038/nmeth.4461 (2017).
Kasuya, G. et al. Cryo-em structures of the human volume-regulated anion channel lrrc8. Nature Structural & Molecular Biology 25, 797–804, https://doi.org/10.1038/s41594-018-0109-6 (2018).
Zivanov, J. et al. New tools for automated high-resolution cryo-em structure determination in relion-3. eLife 7 https://doi.org/10.7554/eLife.42166 (2018).
T, K., N, T. & K, N. First data of beta-galactosidase for validation of the state-of-the-art-cryo em, named cryoarm200 https://doi.org/10.6019/EMPIAR-10204 (2018).
Mei, K. et al. Cryo-em structure of the exocyst complex. Nature Structural & Molecular Biology 25, 139–146, https://doi.org/10.1038/s41594-017-0016-2 (2018).
Danev, R., Yanagisawa, H. & Kikkawa, M. Cryo-electron microscopy methodology: Current aspects and future directions. Trends in Biochemical Sciences 44, 837–848, https://doi.org/10.1016/j.tibs.2019.04.008 (2019).
ET, E. et al. bovine liver glutamate dehydrogenase (18apr21a) https://doi.org/10.6019/EMPIAR-10218 (2018).
Kujirai, T. et al. Structural basis of the nucleosome transition during rna polymerase ii passage. Science 362, 595–598, https://doi.org/10.1126/science.aau9904 (2018).
Dosey, T. L. et al. Structures of trpv2 in distinct conformations provide insight into role of the pore turret. Nature Structural & Molecular Biology 26, 40–49, https://doi.org/10.1038/s41594-018-0168-8 (2018).
Kato, T. et al. Cryotem with a cold field emission gun that moves structural biology into a new stage. Microscopy and Microanalysis 25, 998–999, https://doi.org/10.1017/S1431927619005725 (2019).
Herzik, M. A., Wu, M. & Lander, G. C. High-resolution structure determination of sub-100 kilodalton complexes using conventional cryo-em https://doi.org/10.1101/489898 (2018).
Schmidli, C. et al. Microfluidic protein isolation and sample preparation for high-resolution cryo-em. Proceedings of the National Academy of Sciences 116, 15007–15012, https://doi.org/10.1073/pnas.1907214116 (2019).
Laughlin, T. G., Bayne, A. N., Trempe, J.-F., Savage, D. F. & Davies, K. M. Structure of the complex i-like molecule ndh of oxygenic photosynthesis. Nature 566, 411–414, https://doi.org/10.1038/s41586-019-0921-0 (2019).
Kern, D. M., Oh, S., Hite, R. K. & Brohawn, S. G. Cryo-em structures of the dcpib-inhibited volume-regulated anion channel lrrc8a in lipid nanodiscs. eLife 8, e42636, https://doi.org/10.7554/elife.42636 (2019).
Xu, H. et al. Structural basis of nav1.7 inhibition by a gating-modifier spider toxin. Cell 176, 702–715.e14, https://doi.org/10.1016/j.cell.2018.12.018 (2019).
Li, K. et al. Sub-3 Å apoferritin structure determined with full range of phase shifts using a single position of volta phase plate. Journal of Structural Biology 206, 225–232, https://doi.org/10.1016/j.jsb.2019.03.007 (2019).
Lee, Y. et al. Cryo-em structure of the human l-type amino acid transporter 1 in complex with glycoprotein cd98hc. Nature Structural & Molecular Biology 26, 510–517, https://doi.org/10.1038/s41594-019-0237-7 (2019).
Merchant, M. et al. A bioactive phlebovirus-like envelope protein in a hookworm endogenous retrovirus https://doi.org/10.1101/2021.11.23.469668 (2021).
Fan, X. et al. Single particle cryo-em reconstruction of 52 kda streptavidin at 3.2 angstrom resolution. Nature Communications 10 https://doi.org/10.1038/s41467-019-10368-w (2019).
Sutter, M. et al. Structure of a synthetic β-carboxysome shell. Plant Physiology 181, 1050–1058, https://doi.org/10.1104/pp.19.00885 (2019).
Liu, F. et al. Structural identification of a hotspot on cftr for potentiation. Science 364, 1184–1188, https://doi.org/10.1126/science.aaw7611 (2019).
Ruokolainen, V. et al. Extracellular albumin and endosomal ions prime enterovirus particles for uncoating that can be prevented by fatty acid saturation. Journal of Virology 93 https://doi.org/10.1128/jvi.00599-19 (2019).
Krishna Kumar, K. et al. Structure of a signaling cannabinoid receptor 1-g protein complex. Cell 176, 448–458.e12, https://doi.org/10.1016/j.cell.2018.11.040 (2019).
Tsutsumi, K. et al. Structures of the wild-type mexab–oprm tripartite pump reveal its complex formation and drug efflux mechanism. Nature Communications 10, https://doi.org/10.1038/s41467-019-09463-9 (2019).
Wilkinson, M. E., Kumar, A. & Casañal, A. Methods for merging data sets in electron cryo-microscopy. Acta Crystallographica Section D Structural Biology 75, 782–791, https://doi.org/10.1107/s2059798319010519 (2019).
Hiraizumi, M., Yamashita, K., Nishizawa, T. & Nureki, O. Cryo-em structures capture the transport cycle of the p4-atpase flippase. Science 365, 1149–1155, https://doi.org/10.1126/science.aay3353 (2019).
García-Nafría, J., Nehmé, R., Edwards, P. C. & Tate, C. G. Cryo-em structure of the serotonin 5-ht1b receptor coupled to heterotrimeric go. Nature 558, 620–623, https://doi.org/10.1038/s41586-018-0241-9 (2018).
García-Nafría, J., Lee, Y., Bai, X., Carpenter, B. & Tate, C. G. Cryo-em structure of the adenosine a2a receptor coupled to an engineered heterotrimeric g protein. eLife 7, https://doi.org/10.7554/eLife.35946 (2018).
Suga, M. et al. Structure of the green algal photosystem i supercomplex with a decameric light-harvesting complex i. Nature Plants 5, 626–636, https://doi.org/10.1038/s41477-019-0438-4 (2019).
Hofmann, S. et al. Conformation space of a heterodimeric abc exporter under turnover conditions. Nature 571, 580–583, https://doi.org/10.1038/s41586-019-1391-0 (2019).
Araiso, Y. et al. Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature 575, 395–401, https://doi.org/10.1038/s41586-019-1680-7 (2019).
Ahel, J. et al. Moyamoya disease factor rnf213 is a giant e3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism. eLife 9 https://doi.org/10.7554/elife.56185 (2020).
Wu, M., Lander, G. C. & Herzik, M. A. Sub-2 angstrom resolution structure determination using single-particle cryo-em at 200 kev. Journal of Structural Biology: X 4, 100020, https://doi.org/10.1016/j.yjsbx.2020.100020 (2020).
Lee, Y. et al. Molecular basis of β-arrestin coupling to formoterol-bound β1-adrenoceptor. Nature 583, 862–866, https://doi.org/10.1038/s41586-020-2419-1 (2020).
Kobayashi, K. et al. Cryo-em structure of the human pac1 receptor coupled to an engineered heterotrimeric g protein. Nature Structural & Molecular Biology 27, 274–280, https://doi.org/10.1038/s41594-020-0386-8 (2020).
Coruh, O. et al. Cryo-em structure of a functional monomeric photosystem i from thermosynechococcus elongatus reveals red chlorophyll cluster. Communications Biology 4, 304, https://doi.org/10.1038/s42003-021-01808-9 (2021).
Liang, Y.-L. et al. Toward a structural understanding of class b gpcr peptide binding and activation. Molecular Cell 77, 656–668.e5, https://doi.org/10.1016/j.molcel.2020.01.012 (2020).
Kalbermatter, D. et al. Cryo-em structure of the prefusion state of canine distemper virus fusion protein ectodomain. Journal of Structural Biology: X 4, 100021, https://doi.org/10.1016/j.yjsbx.2020.100021 (2020).
Zhang, Y. et al. Asymmetric opening of the homopentameric 5-ht3a serotonin receptor in lipid bilayers. Nature Communications 12 https://doi.org/10.1038/s41467-021-21016-7 (2021).
Han, H. et al. Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation. Journal of Biological Chemistry 295, 435–443, https://doi.org/10.1074/jbc.AC119.009890 (2020).
Nakamura, R. et al. Cryo-em structure of the volume-regulated anion channel lrrc8d isoform identifies features important for substrate permeation. Communications Biology 3, https://doi.org/10.1038/s42003-020-0951-z (2020).
Bhella, D. Cryo-electron microscopy: an introduction to the technique, and considerations when working to establish a national facility. Biophysical Reviews 11, 515–519, https://doi.org/10.1007/s12551-019-00571-w (2019).
Huang, X. et al. Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation. Progress in Biophysics and Molecular Biology 156, 3–13, https://doi.org/10.1016/j.pbiomolbio.2020.07.009 (2020).
Sears, A. E. et al. Single particle cryo-em of the complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody. The FASEB Journal 34, 13918–13934, https://doi.org/10.1096/fj.202000796rr (2020).
Hurdiss, D. L. et al. Cryo-em structure of coronavirus-hku1 haemagglutinin esterase reveals architectural changes arising from prolonged circulation in humans. Nature Communications 11, https://doi.org/10.1038/s41467-020-18440-6 (2020).
Reid, M. S., Kern, D. M. & Brohawn, S. G. Cryo-em structure of the potassium-chloride cotransporter kcc4 in lipid nanodiscs. eLife 9 https://doi.org/10.7554/eLife.52505. (2020).
Shimada, H. et al. The structure of lipid nanodisc-reconstituted trpv3 reveals the gating mechanism. Nature Structural & Molecular Biology 27, 645–652, https://doi.org/10.1038/s41594-020-0439-z (2020).
Fislage, M., Shkumatov, A. V., Stroobants, A. & Efremov, R. G. Assessing the jeol cryo arm 300 for high-throughput automated single-particle cryo-em in a multiuser environment. IUCrJ 7, 707–718, https://doi.org/10.1107/s2052252520006065 (2020).
Hillen, H. S. et al. Structure of replicating sars-cov-2 polymerase. Nature 584, 154–156, https://doi.org/10.1038/s41586-020-2368-8 (2020).
Park, J. et al. Structure of human gabab receptor in an inactive state. Nature 584, 304–309, https://doi.org/10.1038/s41586-020-2452-0 (2020).
Li, B., Rietmeijer, R. A. & Brohawn, S. G. Structural basis for ph gating of the two-pore domain k+ channel task2. Nature 586, 457–462, https://doi.org/10.1038/s41586-020-2770-2 (2020).
Nakane, T. et al. Single-particle cryo-em at atomic resolution. Nature 587, 152–156, https://doi.org/10.1038/s41586-020-2829-0 (2020).
Burada, A. P., Vinnakota, R. & Kumar, J. Cryo-em structures of the ionotropic glutamate receptor glud1 reveal a non-swapped architecture. Nature Structural & Molecular Biology 27, 84–91, https://doi.org/10.1038/s41594-019-0359-y (2020).
Greber, B. J., Toso, D. B., Fang, J. & Nogales, E. The complete structure of the human tfiih core complex. eLife 8, https://doi.org/10.7554/eLife.44771 (2019).
Greber, B. J. et al. The cryoelectron microscopy structure of the human cdk-activating kinase. Proceedings of the National Academy of Sciences 117, 22849–22857, https://doi.org/10.1073/pnas.2009627117 (2020).
Dijkman, P. M. et al. Structure of the merozoite surface protein 1 from plasmodium falciparum. Science Advances 7, https://doi.org/10.1126/sciadv.abg0465 (2021).
Kern, D. M. et al. Cryo-em structure of the sars-cov-2 3a ion channel in lipid nanodiscs https://doi.org/10.1101/2020.06.17.156554 (2020).
Demura, K. et al. Cryo-em structures of calcium homeostasis modulator channels in diverse oligomeric assemblies. Science Advances 6 https://doi.org/10.1126/sciadv.aba8105 (2020).
Merk, A. et al. 1.8 Å resolution structure of β-galactosidase with a 200 kv cryo arm electron microscope. IUCrJ 7, 639–643, https://doi.org/10.1107/S2052252520006855 (2020).
Qian, H. et al. Inhibition of tetrameric patched1 by sonic hedgehog through an asymmetric paradigm. Nature Communications 10, https://doi.org/10.1038/s41467-019-10234-9 (2019).
Lehmann, L. C. et al. Mechanistic insights into regulation of the alc1 remodeler by the nucleosome acidic patch. Cell Reports 33, 108529, https://doi.org/10.1016/j.celrep.2020.108529 (2020).
Wrobel, A. G. et al. Sars-cov-2 and bat ratg13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nature Structural & Molecular Biology 27, 763–767, https://doi.org/10.1038/s41594-020-0468-7 (2020).
Rzechorzek, N. J., Hardwick, S. W., Jatikusumo, V. A., Chirgadze, D. Y. & Pellegrini, L. Cryoem structures of human cmg–atp γs–dna and cmg–and-1 complexes. Nucleic Acids Research 48, 6980–6995, https://doi.org/10.1093/nar/gkaa429 (2020).
Guo, H. et al. Electron event representation (eer) data enables efficient cryoem file storage with full preservation of spatial and temporal resolution https://doi.org/10.1101/2020.04.28.066795 (2020).
Deng, S., Pan, B., Gottlieb, L., Petersson, E. J. & Marmorstein, R. Molecular basis for n-terminal alpha-synuclein acetylation by human natb https://doi.org/10.1101/2020.05.11.089318. (2020).
Flores, J. A. et al. Connexin-46/50 in a dynamic lipid environment resolved by cryoem at 1.9 Å. Nature Communications 11, https://doi.org/10.1038/s41467-020-18120-5 (2020).
Syrjanen, J. L. et al. Structure and assembly of calcium homeostasis modulator proteins. Nature Structural & Molecular Biology 27, 150–159, https://doi.org/10.1038/s41594-019-0369-9 (2020).
Ke, Z. et al. Structures and distributions of sars-cov-2 spike proteins on intact virions. Nature 588, 498–502, https://doi.org/10.1038/s41586-020-2665-2 (2020).
Reddy, B., Bavi, N., Lu, A., Park, Y. & Perozo, E. Molecular basis of force-from-lipids gating in the mechanosensitive channel mscs. eLife 8 https://doi.org/10.7554/elife.50486 (2019).
He, S. et al. The structural basis of rubisco phase separation in the pyrenoid. Nature Plants 6, 1480–1490, https://doi.org/10.1038/s41477-020-00811-y (2020).
Nichols, R. J. et al. Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism https://doi.org/10.1101/2020.05.24.113720 (2020).
Tsutsumi, N. et al. Structure of human frizzled5 by fiducial-assisted cryo-em supports a heterodimeric mechanism of canonical wnt signaling. eLife 9 https://doi.org/10.7554/eLife.58464 (2020).
Shams, A. et al. Comprehensive deletion landscape of crispr-cas9 identifies minimal rna-guided dna-binding modules. Nature Communications 12 https://doi.org/10.1038/s41467-021-25992-8 (2021).
Watson, Z. L. et al. Structure of the bacterial ribosome at 2 Å resolution. eLife 9, https://doi.org/10.7554/elife.60482 (2020).
Zhao, B. et al. The molecular basis of tight nuclear tethering and inactivation of cgas. Nature 587, 673–677, https://doi.org/10.1038/s41586-020-2749-z (2020).
Vizarraga, D. et al. Immunodominant proteins p1 and p40/p90 from human pathogen mycoplasma pneumoniae. Nature Communications 11 https://doi.org/10.1038/s41467-020-18777-y (2020).
Li, Q. et al. Synthetic group a streptogramin antibiotics that overcome vat resistance. Nature 586, 145–150, https://doi.org/10.1038/s41586-020-2761-3 (2020).
Capodagli, G. C. et al. Structure–function studies of rgg binding to pheromones and target promoters reveal a model of transcription factor interplay. Proceedings of the National Academy of Sciences 117, 24494–24502, https://doi.org/10.1073/pnas.2008427117 (2020).
Coudray, N. et al. Structure of bacterial phospholipid transporter mlafedb with substrate bound. eLife 9, https://doi.org/10.7554/elife.62518 (2020).
Crowe-McAuliffe, C. et al. Structural basis for bacterial ribosome-associated quality control by rqch and rqcp. Molecular Cell 81, 115–126.e7, https://doi.org/10.1016/j.molcel.2020.11.002 (2021).
Madej, M. et al. Structural and functional insights into oligopeptide acquisition by the ragab transporter from porphyromonas gingivalis. Nature Microbiology 5, 1016–1025, https://doi.org/10.1038/s41564-020-0716-y (2020).
Kondo, Y. et al. Cryo-em structure of a dimeric b-raf:14-3-3 complex reveals asymmetry in the active sites of b-raf kinases. Science 366, 109–115, https://doi.org/10.1126/science.aay0543 (2019).
Sato, Y. et al. Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase from sulfurisphaera tokodaii. Journal of Structural Biology: X 4, 100030, https://doi.org/10.1016/j.yjsbx.2020.100030 (2020).
Feathers, J. R., Spoth, K. A. & Fromme, J. C. Experimental evaluation of super-resolution imaging and magnification choice in single-particle cryo-em. Journal of Structural Biology: X 5, 100047, https://doi.org/10.1016/j.yjsbx.2021.100047 (2021).
Velazhahan, V. et al. Structure of the class d gpcr ste2 dimer coupled to two g proteins. Nature 589, 148–153, https://doi.org/10.1038/s41586-020-2994-1 (2020).
Purushotham, P., Ho, R. & Zimmer, J. Architecture of a catalytically active homotrimeric plant cellulose synthase complex. Science 369, 1089–1094, https://doi.org/10.1126/science.abb2978 (2020).
Kato, K. et al. High-resolution cryo-em structure of photosystem ii: Effects of electron beam damage https://doi.org/10.1101/2020.10.18.344648 (2020).
Saxton, R. A. et al. Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10. Science 371, https://doi.org/10.1126/science.abc8433 (2021).
Caffalette, C. A. & Zimmer, J. Cryo-em structure of the full-length wzmwzt abc transporter required for lipid-linked o antigen transport. Proceedings of the National Academy of Sciences 118, https://doi.org/10.1073/pnas.2016144118 (2020).
Danev, R., Yanagisawa, H. & Kikkawa, M. Cryo-em performance testing of hardware and data acquisition strategies. Microscopy 70, 487–497, https://doi.org/10.1093/jmicro/dfab016 (2021).
Greber, B. J., Remis, J., Ali, S. & Nogales, E. 2.5 Å-resolution structure of human cdk-activating kinase bound to the clinical inhibitor icec0942. Biophysical Journal 120, 677–686, https://doi.org/10.1016/j.bpj.2020.12.030 (2021).
Ariyoshi, M. et al. Cryo-em structure of the cenp-a nucleosome in complex with phosphorylated cenp-c. The EMBO Journal 40, https://doi.org/10.15252/embj.2020105671 (2021).
Robert Hollingsworth, L. et al. Mechanism of filament formation in upa-promoted card8 and nlrp1 inflammasomes. Nature Communications 12, https://doi.org/10.1038/s41467-020-20320-y (2021).
Zhou, B.-R. et al. Distinct structures and dynamics of chromatosomes with different human linker histone isoforms. Molecular Cell 81, 166–182.e6, https://doi.org/10.1016/j.molcel.2020.10.038 (2021).
Lees, J. A., Li, P., Kumar, N., Weisman, L. S. & Reinisch, K. M. Insights into lysosomal pi(3,5)p2 homeostasis from a structural-biochemical analysis of the pikfyve lipid kinase complex. Molecular Cell 80, 736–743.e4, https://doi.org/10.1016/j.molcel.2020.10.003 (2020).
Adachi, N. et al. Sub-3 Å resolution structure of 110 kda nitrite reductase determined by 200 kv cryogenic electron microscopy https://doi.org/10.1101/2020.07.12.199695 (2020).
Niu, Y., Suzuki, H., Hosford, C. J., Walz, T. & Chappie, J. S. Structural asymmetry governs the assembly and gtpase activity of mcrbc restriction complexes. Nature Communications 11, https://doi.org/10.1038/s41467-020-19735-4 (2020).
Takeda, S. N. et al. Structure of the miniature type v-f crispr-cas effector enzyme. Molecular Cell 81, 558–570.e3, https://doi.org/10.1016/j.molcel.2020.11.035 (2021).
Maldonado, M., Padavannil, A., Zhou, L., Guo, F. & Letts, J. A. Atomic structure of a mitochondrial complex i intermediate from vascular plants. eLife 9, https://doi.org/10.7554/elife.56664 (2020).
Sun, M. et al. Practical considerations for using k3 cameras in cds mode for high-resolution and high-throughput single particle cryo-em. Journal of Structural Biology 213, 107745, https://doi.org/10.1016/j.jsb.2021.107745 (2021).
Yip, K. M., Fischer, N., Paknia, E., Chari, A. & Stark, H. Atomic-resolution protein structure determination by cryo-em. Nature 587, 157–161, https://doi.org/10.1038/s41586-020-2833-4 (2020).
Hollingsworth, L. R. et al. Dpp9 sequesters the c terminus of nlrp1 to repress inflammasome activation. Nature 592, 778–783, https://doi.org/10.1038/s41586-021-03350-4 (2021).
Sharif, H. et al. Dipeptidyl peptidase 9 sets a threshold for card8 inflammasome formation by sequestering its active c-terminal fragment. Immunity 54, 1392–1404.e10, https://doi.org/10.1016/j.immuni.2021.04.024 (2021).
Hsia, Y. et al. Design of multi-scale protein complexes by hierarchical building block fusion. Nature Communications 12, 2294, https://doi.org/10.1038/s41467-021-22276-z (2021).
Yin, Z. et al. Structural basis for a complex i mutation that blocks pathological ros production. Nature Communications 12, 707, https://doi.org/10.1038/s41467-021-20942-w (2021).
Zhang, K. et al. Cryo-em structures of helicobacter pylori vacuolating cytotoxin a oligomeric assemblies at near-atomic resolution. Proceedings of the National Academy of Sciences 116, 6800–6805, https://doi.org/10.1073/pnas.1821959116 (2019).
Shakeel, S. et al. Structure of the fanconi anaemia monoubiquitin ligase complex. Nature 575, 234–237, https://doi.org/10.1038/s41586-019-1703-4 (2019).
Alcón, P. et al. Fancd2–fanci is a clamp stabilized on dna by monoubiquitination of fancd2 during dna repair. Nature Structural & Molecular Biology 27, 240–248, https://doi.org/10.1038/s41594-020-0380-1 (2020).
Acheson, J. F., Ho, R., Goularte, N. F., Cegelski, L. & Zimmer, J. Molecular organization of the e. coli cellulose synthase macrocomplex. Nature Structural & Molecular Biology 28, 310–318, https://doi.org/10.1038/s41594-021-00569-7 (2021).
Asai, T. et al. Cryo-em structure of k+-bound herg channel complexed with the blocker astemizole. Structure 29, 203–212.e4, https://doi.org/10.1016/j.str.2020.12.007 (2021).
Zhang, K., Pintilie, G. D., Li, S., Schmid, M. F. & Chiu, W. Resolving individual atoms of protein complex by cryo-electron microscopy. Cell Research 30, 1136–1139, https://doi.org/10.1038/s41422-020-00432-2 (2020).
Robinson, R. A. et al. Simultaneous binding of guidance cues net1 and rgm blocks extracellular neo1 signaling. Cell 184, 2103–2120.e31, https://doi.org/10.1016/j.cell.2021.02.045 (2021).
RG, E. & AV, S. Coma-corrected rapid single-particle cryo-em data collection on the cryoarm300 https://doi.org/10.6019/EMPIAR-10639 (2021).
M, H., K, Y., T, N., M, K. & O, N. 1.93 a cryo-em structure of streptavidin https://doi.org/10.6019/EMPIAR-10641 (2021).
Wang, J. Y. et al. Structural coordination between active sites of a cas6-reverse transcriptase-cas1-cas2 crispr integrase complex https://doi.org/10.1101/2020.10.18.344481 (2020).
Saur, M. et al. Fragment-based drug discovery using cryo-em. Drug Discovery Today 25, 485–490, https://doi.org/10.1016/j.drudis.2019.12.006 (2020).
Khanra, N., Brown, P. M., Perozzo, A. M., Bowie, D. & Meyerson, J. R. Architecture and structural dynamics of the heteromeric gluk2/k5 kainate receptor. eLife 10 https://doi.org/10.7554/elife.66097 (2021).
Gunasekar, S. K. et al. Small molecule swell1-lrrc8 complex induction improves glycemic control and nonalcoholic fatty liver disease in murine type 2 diabetes https://doi.org/10.1101/2021.02.28.432901 (2021).
Lo, W.-T. et al. Structural basis of phosphatidylinositol 3-kinase c2α function. Nature Structural & Molecular Biology 29, 218–228, https://doi.org/10.1038/s41594-022-00730-w (2022).
Coscia, F. et al. The structure of human thyroglobulin. Nature 578, 627–630, https://doi.org/10.1038/s41586-020-1995-4 (2020).
Josephs, T. M. et al. Structure and dynamics of the cgrp receptor in apo and peptide-bound forms. Science 372 https://doi.org/10.1126/science.abf7258 (2021).
Zhang, X. et al. Differential glp-1r binding and activation by peptide and non-peptide agonists. Molecular Cell 80, 485–500.e7, https://doi.org/10.1016/j.molcel.2020.09.020 (2020).
Wiryaman, T. & Toor, N. Cryo-em structure of a thermostable bacterial nanocompartment. IUCrJ 8, 342–350, https://doi.org/10.1107/s2052252521001949 (2021).
Gupta, T. K. et al. Structural basis for vipp1 oligomerization and maintenance of thylakoid membrane integrity. Cell 184, 3643–3659.e23, https://doi.org/10.1016/j.cell.2021.05.011 (2021).
Crowe-McAuliffe, C. et al. Structural basis of abcf-mediated resistance to pleuromutilin, lincosamide, and streptogramin a antibiotics in gram-positive pathogens. Nature Communications 12, https://doi.org/10.1038/s41467-021-23753-1 (2021).
Arimura, Y., Shih, R. M., Froom, R. & Funabiki, H. Structural features of nucleosomes in interphase and metaphase chromosomes. Molecular Cell 81, 4377–4397.e12, https://doi.org/10.1016/j.molcel.2021.08.010 (2021).
van der Stel, A.-X. et al. Structural basis for the tryptophan sensitivity of tnac-mediated ribosome stalling. Nature Communications 12, https://doi.org/10.1038/s41467-021-25663-8 (2021).
Girbig, M. et al. Cryo-em structures of human rna polymerase iii in its unbound and transcribing states. Nature Structural & Molecular Biology 28, 210–219, https://doi.org/10.1038/s41594-020-00555-5 (2021).
Cater, R. J. et al. Structural basis of omega-3 fatty acid transport across the blood–brain barrier. Nature 595, 315–319, https://doi.org/10.1038/s41586-021-03650-9 (2021).
Nygaard, R. et al. Structural basis of wls/evi-mediated wnt transport and secretion. Cell 184, 194–206.e14, https://doi.org/10.1016/j.cell.2020.11.038 (2021).
Huber, S. T. et al. Nanofluidic chips for cryo-em structure determination from picoliter sample volumes. eLife 11, https://doi.org/10.7554/elife.72629 (2022).
de Martín Garrido, N. et al. Structure of the bacteriophage phikz non-virion rna polymerase. Nucleic Acids Research 49, 7732–7739, https://doi.org/10.1093/nar/gkab539 (2021).
J, A. et al. E3 ubiquitin ligase rnf213 employs a non-canonical zinc finger active site and is allosterically regulated by atp https://doi.org/10.6019/EMPIAR-10711 (2021).
J, A. et al. E3 ubiquitin ligase rnf213 employs a non-canonical zinc finger active site and is allosterically regulated by atp https://doi.org/10.6019/EMPIAR-10712 (2020).
Okamoto, H. H. et al. Cryo-em structure of the human mt1–gi signaling complex. Nature Structural & Molecular Biology 28, 694–701, https://doi.org/10.1038/s41594-021-00634-1 (2021).
Ramlaul, K. et al. Architecture of the tuberous sclerosis protein complex. Journal of Molecular Biology 433, 166743, https://doi.org/10.1016/j.jmb.2020.166743 (2021).
Ross, J. et al. Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment. Science Advances 8, https://doi.org/10.1126/sciadv.abj4461 (2022).
Prattes, M. et al. Structural basis for inhibition of the aaa-atpase drg1 by diazaborine. Nature Communications 12, https://doi.org/10.1038/s41467-021-23854-x (2021).
D, M. & C, S. structure of human haemoglobin obtained via cryoelectron microscopy at 200 kv https://doi.org/10.6019/EMPIAR-10721 (2021).
Zhou, T. et al. Cryo-em structures of sars-cov-2 spike without and with ace2 reveal a ph-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host & Microbe 28, 867–879.e5, https://doi.org/10.1016/j.chom.2020.11.004 (2020).
Benton, D. J. et al. The effect of the d614g substitution on the structure of the spike glycoprotein of sars-cov-2. Proceedings of the National Academy of Sciences 118, https://doi.org/10.1073/pnas.2022586118 (2021).
Takada, H. et al. Rqch and rqcp catalyze processive poly-alanine synthesis in a reconstituted ribosome-associated quality control system. Nucleic Acids Research 49, 8355–8369, https://doi.org/10.1093/nar/gkab589 (2021).
Sauer, D. B. et al. Structure and inhibition mechanism of the human citrate transporter nact. Nature 591, 157–161, https://doi.org/10.1038/s41586-021-03230-x (2021).
Ghanim, G. E. et al. Structure of human telomerase holoenzyme with bound telomeric dna. Nature 593, 449–453, https://doi.org/10.1038/s41586-021-03415-4 (2021).
Fujita-Fujiharu, Y. et al. Structural insight into marburg virus nucleoprotein–rna complex formation. Nature Communications 13, https://doi.org/10.1038/s41467-022-28802-x (2022).
Xiong, X. et al. Symmetric and asymmetric receptor conformation continuum induced by a new insulin. Nature Chemical Biology 18, 511–519, https://doi.org/10.1038/s41589-022-00981-0 (2022).
Li, J. et al. Cryo-em structures of escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site. Proceedings of the National Academy of Sciences 118, https://doi.org/10.1073/pnas.2106750118 (2021).
Bacic, L. et al. Structure and dynamics of the chromatin remodeler alc1 bound to a parylated nucleosome. eLife 10, https://doi.org/10.7554/elife.71420 (2021).
Jochheim, F. A. et al. The structure of a dimeric form of sars-cov-2 polymerase. Communications Biology 4, https://doi.org/10.1038/s42003-021-02529-9 (2021).
Baker, A. T. et al. The structure of chadox1/azd-1222 reveals interactions with car and pf4 with implications for vaccine-induced immune thrombotic thrombocytopenia https://doi.org/10.1101/2021.05.19.444882 (2021).
Kuzuya, M. et al. Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the n terminus and phospholipids. Science Signaling 15, https://doi.org/10.1126/scisignal.abg6941 (2022).
Ghilarov, D. et al. Molecular mechanism of sbma, a promiscuous transporter exploited by antimicrobial peptides. Science Advances 7 https://doi.org/10.1126/sciadv.abj5363 (2021).
Crowe-McAuliffe, C. et al. Structural basis for poxta-mediated resistance to phenicol and oxazolidinone antibiotics https://doi.org/10.1101/2021.06.18.448924 (2021).
Pöll, G., Pilsl, M., Griesenbeck, J., Tschochner, H. & Milkereit, P. Analysis of subunit folding contribution of three yeast large ribosomal subunit proteins required for stabilisation and processing of intermediate nuclear rrna precursors https://doi.org/10.1101/2021.05.18.444632 (2021).
Melville, Z., Kim, K., Clarke, O. B. & Marks, A. R. High resolution structure of the membrane embedded skeletal muscle ryanodine receptor https://doi.org/10.1101/2021.03.09.434632 (2021).
Cook, A. D., Manka, S. W., Wang, S., Moores, C. A. & Atherton, J. A microtubule relion-based pipeline for cryo-em image processing. Journal of Structural Biology 209, 107402, https://doi.org/10.1016/j.jsb.2019.10.004 (2020).
Johnson, Z. L. & Chen, J. Structural basis of substrate recognition by the multidrug resistance protein mrp1. Cell 168, 1075–1085.e9, https://doi.org/10.1016/j.cell.2017.01.041 (2017).
Kim, Y. & Chen, J. Molecular structure of human p-glycoprotein in the atp-bound, outward-facing conformation. Science 359, 915–919, https://doi.org/10.1126/science.aar7389 (2018).
Conners, R. et al. Cryoem structure of the outer membrane secretin channel piv from the f1 filamentous bacteriophage. Nature Communications 12 https://doi.org/10.1038/s41467-021-26610-3 (2021).
Liu, F., Lee, J. & Chen, J. Molecular structures of the eukaryotic retinal importer abca4. eLife 10, https://doi.org/10.7554/elife.63524 (2021).
Desai, N. et al. Elongational stalling activates mitoribosome-associated quality control. Science 370, 1105–1110, https://doi.org/10.1126/science.abc7782 (2020).
AS, G. Cryo-em structure of sars-cov-2 main protease c145s in complex with n-terminal peptide https://doi.org/10.6019/EMPIAR-10810 (2021).
Oldham, M. L., Grigorieff, N. & Chen, J. Structure of the transporter associated with antigen processing trapped by herpes simplex virus. eLife 5, https://doi.org/10.7554/elife.21829 (2016).
A, M., JE, D., R, G. & J, O. 2.1 Å resolution structure of β-galactosidase obtained from glacios equipped with falcon 3 https://doi.org/10.6019/EMPIAR-10817 (2021).
Vieni, C., Coudray, N., Isom, G. L., Bhabha, G. & Ekiert, D. C. Role of ring6 in the function of the e. coli mce protein letb. Journal of Molecular Biology 434, 167463, https://doi.org/10.1016/j.jmb.2022.167463 (2022).
Lancey, C. et al. Structure of the processive human pol δ holoenzyme. Nature Communications 11, https://doi.org/10.1038/s41467-020-14898-6 (2020).
Wälti, M. A., Canagarajah, B., Schwieters, C. D. & Clore, G. M. Visualization of sparsely-populated lower-order oligomeric states of human mitochondrial hsp60 by cryo-electron microscopy. Journal of Molecular Biology 433, 167322, https://doi.org/10.1016/j.jmb.2021.167322 (2021).
Chen, M. et al. Molecular architecture of black widow spider neurotoxins. Nature Communications 12, https://doi.org/10.1038/s41467-021-26562-8 (2021).
Kim, K. et al. The structure of natively iodinated bovine thyroglobulin. Acta Crystallographica Section D Structural Biology 77, 1451–1459, https://doi.org/10.1107/s2059798321010056 (2021).
Park, J. et al. Symmetric activation and modulation of the human calcium-sensing receptor. Proceedings of the National Academy of Sciences 118, https://doi.org/10.1073/pnas.2115849118 (2021).
Horne, C. R. et al. Mechanism of nanr gene repression and allosteric induction of bacterial sialic acid metabolism. Nature Communications 12, https://doi.org/10.1038/s41467-021-22253-6 (2021).
Sun, D. et al. Potent neutralizing nanobodies resist convergent circulating variants of sars-cov-2 by targeting diverse and conserved epitopes. Nature Communications 12, https://doi.org/10.1038/s41467-021-24963-3 (2021).
Li, B., Hoel, C. M. & Brohawn, S. G. Structures of tweety homolog proteins ttyh2 and ttyh3 reveal a Ca2+-dependent switch from intra- to inter-membrane dimerization https://doi.org/10.1101/2021.08.15.456437 (2021).
D, K., L, Y. & A, K. Cryoem structure of apoferritin at 2.6a from tundra, 100kv microscope https://doi.org/10.6019/EMPIAR-10844 (2021).
Webster, M. W. et al. Structural basis of transcription-translation coupling and collision in bacteria. Science 369, 1355–1359, https://doi.org/10.1126/science.abb5036 (2020).
Sae-Lee, W. et al. The protein organization of a red blood cell. Cell Reports 40, 111103, https://doi.org/10.1016/j.celrep.2022.111103 (2022).
Basanta, B., Hirschi, M. M., Grotjahn, D. A. & Lander, G. C. A case for glycerol as an acceptable additive for single particle cryoem samples https://doi.org/10.1101/2021.09.10.459874 (2021).
Lilic, M., Darst, S. A. & Campbell, E. A. Structural basis of transcriptional activation by the mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor whib7. Molecular Cell 81, 2875–2886.e5, https://doi.org/10.1016/j.molcel.2021.05.017 (2021).
Cao, C. et al. Structure, function and pharmacology of human itch gpcrs. Nature 600, 170–175, https://doi.org/10.1038/s41586-021-04126-6 (2021).
D, K., AF, K., L, Y. & A, K. Cryoem structure of gaba(a)r-beta3 homopentamer at 3.4a from tundra, 100kv microscope https://doi.org/10.6019/EMPIAR-10858 (2021).
Laughlin, T. G. et al. Architecture and self-assembly of the jumbo bacteriophage nuclear shell https://doi.org/10.1101/2022.02.14.480162 (2022).
Yang, C. et al. Structural visualization of de novo transcription initiation by saccharomyces cerevisiae rna polymerase ii. Molecular Cell 82, 660–676.e9, https://doi.org/10.1016/j.molcel.2021.12.020 (2022).
Li, Y. et al. Oligomeric interactions maintain active-site structure in a noncooperative enzyme family. The EMBO Journal 41, https://doi.org/10.15252/embj.2021108368 (2022).
Donovan, B. T. et al. Basic helix-loop-helix pioneer factors interact with the histone octamer to invade nucleosomes and generate nucleosome-depleted regions. Molecular Cell 83, 1251–1263.e6, https://doi.org/10.1016/j.molcel.2023.03.006 (2023).
Velazhahan, V., Ma, N., Vaidehi, N. & Tate, C. G. Activation mechanism of the class d fungal gpcr dimer ste2. Nature 603, 743–748, https://doi.org/10.1038/s41586-022-04498-3 (2022).
Qiao, C., Debiasi-Anders, G. & Mir-Sanchis, I. Staphylococcal self-loading helicases couple the staircase mechanism with inter domain high flexibility. Nucleic Acids Research 50, 8349–8362, https://doi.org/10.1093/nar/gkac625 (2022).
Pan, M. et al. Structural insights into ubr1-mediated n-degron polyubiquitination. Nature 600, 334–338, https://doi.org/10.1038/s41586-021-04097-8 (2021).
Boyaci, H. et al. Fidaxomicin jams mycobacterium tuberculosis rna polymerase motions needed for initiation via rbpa contacts. eLife 7, https://doi.org/10.7554/elife.34823 (2018).
Skalidis, I. et al. Cryo-em and artificial intelligence visualize endogenous protein community members. Structure 30, 575–589.e6, https://doi.org/10.1016/j.str.2022.01.001 (2022).
Moghadamchargari, Z. et al. Molecular assemblies of the catalytic domain of sos with kras and oncogenic mutants. Proceedings of the National Academy of Sciences 118, https://doi.org/10.1073/pnas.2022403118 (2021).
Boyaci, H., Chen, J., Jansen, R., Darst, S. A. & Campbell, E. A. Structures of an rna polymerase promoter melting intermediate elucidate dna unwinding. Nature 565, 382–385, https://doi.org/10.1038/s41586-018-0840-5 (2019).
Sente, A. et al. Differential assembly diversifies gabaa receptor structures and signalling. Nature 604, 190–194, https://doi.org/10.1038/s41586-022-04517-3 (2022).
Fresquet, M. et al. Structure of pla2r reveals presentation of the dominant membranous nephropathy epitope and an immunogenic patch. Proceedings of the National Academy of Sciences 119, https://doi.org/10.1073/pnas.2202209119 (2022).
Snead, D. M. et al. Structural basis for parkinson’s disease-linked lrrk2’s binding to microtubules. Nature Structural & Molecular Biology 29, 1196–1207, https://doi.org/10.1038/s41594-022-00863-y (2022).
Kishi, K. E. et al. Structural basis for channel conduction in the pump-like channelrhodopsin chrmine. Cell 185, 672–689.e23, https://doi.org/10.1016/j.cell.2022.01.007 (2022).
Grba, D. N. et al. Cryo-electron microscopy reveals how acetogenins inhibit mitochondrial respiratory complex i. Journal of Biological Chemistry 298, 101602, https://doi.org/10.1016/j.jbc.2022.101602 (2022).
Li, J. et al. Structure of cyanobacterial photosystem i complexed with ferredoxin at 1.97 Å resolution. Communications Biology 5, https://doi.org/10.1038/s42003-022-03926-4 (2022).
Robertson, M. J., Meyerowitz, J. G., Panova, O., Borrelli, K. & Skiniotis, G. Plasticity in ligand recognition at somatostatin receptors. Nature Structural & Molecular Biology 29, 210–217, https://doi.org/10.1038/s41594-022-00727-5 (2022).
Meyerowitz, J. G. et al. The oxytocin signaling complex reveals a molecular switch for cation dependence. Nature Structural & Molecular Biology 29, 274–281, https://doi.org/10.1038/s41594-022-00728-4 (2022).
Markert, J., Zhou, K. & Luger, K. Smarcad1 is an atp-dependent histone octamer exchange factor with de novo nucleosome assembly activity. Science Advances 7, https://doi.org/10.1126/sciadv.abk2380 (2021).
Thangaratnarajah, C., Rheinberger, J., Paulino, C. & Slotboom, D. J. Insights into the bilayer-mediated toppling mechanism of a folate-specific ecf transporter by cryo-em. Proceedings of the National Academy of Sciences 118, https://doi.org/10.1073/pnas.2105014118 (2021).
Fujita, J. et al. Epoxidized graphene grid for highly efficient high-resolution cryoem structural analysis. Scientific Reports 13, https://doi.org/10.1038/s41598-023-29396-0 (2023).
D, K., AF, K., L, Y. & A, K. Cryoem structure of t20s proteasome at 3a from tundra, 100kv microscope https://doi.org/10.6019/EMPIAR-10961 (2022).
Baudin, F. et al. Mechanism of rna polymerase i selection by transcription factor uaf. Science Advances 8, https://doi.org/10.1126/sciadv.abn5725 (2022).
Misiaszek, A. D. et al. Cryo-em structures of human rna polymerase i. Nature Structural & Molecular Biology 28, 997–1008, https://doi.org/10.1038/s41594-021-00693-4 (2021).
Kandolf, S. et al. Cryo-em structure of the plant 26s proteasome. Plant Communications 3, 100310, https://doi.org/10.1016/j.xplc.2022.100310 (2022).
Tomita, A. et al. Cryo-em reveals mechanistic insights into lipid-facilitated polyamine export by human atp13a2. Molecular Cell 81, 4799–4809.e5, https://doi.org/10.1016/j.molcel.2021.11.001 (2021).
FA, K., K, K. & A, K. 2.1Å t20s proteosome from 200kv glacios with selectris falcon 4 https://doi.org/10.6019/EMPIAR-10976 (2022).
Seven, A. B. et al. G-protein activation by a metabotropic glutamate receptor. Nature 595, 450–454, https://doi.org/10.1038/s41586-021-03680-3 (2021).
Lubbe, L., Sewell, B. T., Woodward, J. D. & Sturrock, E. D. Cryo-em reveals mechanisms of angiotensin i-converting enzyme allostery and dimerization. The EMBO Journal 41, https://doi.org/10.15252/embj.2021110550 (2022).
Bridges, H. R. et al. Structural basis of mammalian respiratory complex i inhibition by medicinal biguanides. Science 379, 351–357, https://doi.org/10.1126/science.ade3332 (2023).
Milazzo, F. M. et al. Spike mutation resilient scfv76 antibody counteracts sars-cov-2 lung damage upon aerosol delivery. Molecular Therapy 31, 362–373, https://doi.org/10.1016/j.ymthe.2022.09.010 (2023).
Arragain, B. et al. Structural snapshots of la crosse virus polymerase reveal the mechanisms underlying peribunyaviridae replication and transcription. Nature Communications 13, https://doi.org/10.1038/s41467-022-28428-z (2022).
K, K. et al. Endogenous ligand recognition and structural transition of a human pth receptor. https://doi.org/10.6019/EMPIAR-10996 (2022).
Z, M. et al. A drug and atp binding site in type 1 ryanodine receptor https://doi.org/10.6019/EMPIAR-10997 (2022).
Yang, K. et al. Structural conservation among variants of the sars-cov-2 spike postfusion bundle. Proceedings of the National Academy of Sciences 119, https://doi.org/10.1073/pnas.2119467119 (2022).
Tholen, J., Razew, M., Weis, F. & Galej, W. P. Structural basis of branch site recognition by the human spliceosome. Science 375, 50–57, https://doi.org/10.1126/science.abm4245 (2022).
Fiedorczuk, K. & Chen, J. Mechanism of cftr correction by type i folding correctors. Cell 185, 158–168.e11, https://doi.org/10.1016/j.cell.2021.12.009 (2022).
Kieuvongngam, V. & Chen, J. Structures of the peptidase-containing abc transporter pcat1 under equilibrium and nonequilibrium conditions. Proceedings of the National Academy of Sciences 119, https://doi.org/10.1073/pnas.2120534119 (2022).
Basu, R. S., Sherman, M. B. & Gagnon, M. G. Compact if2 allows initiator trna accommodation into the p site and gates the ribosome to elongation. Nature Communications 13, https://doi.org/10.1038/s41467-022-31129-2 (2022).
Brown, H. G. & Hanssen, E. Measureice: accessible on-the-fly measurement of ice thickness in cryo-electron microscopy. Communications Biology 5, https://doi.org/10.1038/s42003-022-03698-x (2022).
Pulkkinen, L. I. A. et al. Molecular organisation of tick-borne encephalitis virus. Viruses 14, 792, https://doi.org/10.3390/v14040792 (2022).
Rangarajan, E. S. & Izard, T. The cryogenic electron microscopy structure of the cell adhesion regulator metavinculin reveals an isoform-specific kinked helix in its cytoskeleton binding domain. International Journal of Molecular Sciences 22, 645, https://doi.org/10.3390/ijms22020645 (2021).
Maloney, F. P. et al. Structure, substrate recognition and initiation of hyaluronan synthase. Nature 604, 195–201, https://doi.org/10.1038/s41586-022-04534-2 (2022).
CM, H., L, Z. & SG, B. Cryo-em structure of the gold-domain seven-transmembrane protein tmem87a https://doi.org/10.6019/EMPIAR-11045 (2022).
Sakuragi, T. et al. The tertiary structure of the human xkr8–basigin complex that scrambles phospholipids at plasma membranes. Nature Structural & Molecular Biology 28, 825–834, https://doi.org/10.1038/s41594-021-00665-8 (2021).
Zhao, Y. et al. Structure of the human cation–chloride cotransport kcc1 in an outward-open state. Proceedings of the National Academy of Sciences 119, https://doi.org/10.1073/pnas.2109083119 (2022).
Zhao, Y. et al. Structural basis for inhibition of the cation-chloride cotransporter nkcc1 by the diuretic drug bumetanide. Nature Communications 13, https://doi.org/10.1038/s41467-022-30407-3 (2022).
Newing, T. P. et al. Molecular basis for rna polymerase-dependent transcription complex recycling by the helicase-like motor protein held. Nature Communications 11, https://doi.org/10.1038/s41467-020-20157-5 (2020).
Prattes, M. et al. Visualizing maturation factor extraction from the nascent ribosome by the aaa-atpase drg1. Nature Structural & Molecular Biology 29, 942–953, https://doi.org/10.1038/s41594-022-00832-5 (2022).
Clark, M. D., Contreras, G. F., Shen, R. & Perozo, E. Electromechanical coupling in the hyperpolarization-activated k+ channel kat1. Nature 583, 145–149, https://doi.org/10.1038/s41586-020-2335-4 (2020).
Asami, J. et al. Structure of the bile acid transporter and hbv receptor ntcp. Nature 606, 1021–1026, https://doi.org/10.1038/s41586-022-04845-4 (2022).
Tanaka, S. et al. Structural basis for binding of potassium-competitive acid blockers to the gastric proton pump. Journal of Medicinal Chemistry 65, 7843–7853, https://doi.org/10.1021/acs.jmedchem.2c00338 (2022).
Katsuyama, Y. et al. Structural and functional analyses of the tridomain-nonribosomal peptide synthetase fmoa3 for 4-methyloxazoline ring formation. Angewandte Chemie International Edition 60, 14554–14562, https://doi.org/10.1002/anie.202102760 (2021).
Oki, K. et al. Dna polymerase d temporarily connects primase to the cmg-like helicase before interacting with proliferating cell nuclear antigen. Nucleic Acids Research 49, 4599–4612, https://doi.org/10.1093/nar/gkab243 (2021).
Day, M., Oliver, A. W. & Pearl, L. H. Structure of the human rad17-rfc clamp loader and 9-1-1 checkpoint clamp bound to a dsdna-ssdna junction https://doi.org/10.1101/2022.03.11.484023 (2022).
Dolan, K. A. et al. Structure of sars-cov-2 m protein in lipid nanodiscs https://doi.org/10.1101/2022.06.12.495841 (2022).
Morreale, F. E. et al. Bacprotacs mediate targeted protein degradation in bacteria. Cell 185, 2338–2353.e18, https://doi.org/10.1016/j.cell.2022.05.009 (2022).
Schmidt, L. et al. Delineating organizational principles of the endogenous l-a virus by cryo-em and computational analysis of native cell extracts https://doi.org/10.1101/2022.07.15.498668 (2022).
Selvakumar, P. et al. Structures of the t cell potassium channel kv1.3 with immunoglobulin modulators. Nature Communications 13, https://doi.org/10.1038/s41467-022-31285-5 (2022).
Teramoto, T. et al. Minimal protein-only rnase p structure reveals insights into trna precursor recognition and catalysis. Journal of Biological Chemistry 297, 101028, https://doi.org/10.1016/j.jbc.2021.101028 (2021).
Suzuki, S. et al. Structural insight into the activation mechanism of mrgd with heterotrimeric gi-protein revealed by cryo-em. Communications Biology 5, https://doi.org/10.1038/s42003-022-03668-3 (2022).
S, S. et al. Structural insight into the activation mechanism of mrgd with heterotrimeric gi-protein revealed by cryo-em https://doi.org/10.6019/EMPIAR-11074 (2022).
Barandun, J., Hunziker, M., Vossbrinck, C. R. & Klinge, S. Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome. Nature Microbiology 4, 1798–1804, https://doi.org/10.1038/s41564-019-0514-6 (2019).
Ehrenbolger, K. et al. Differences in structure and hibernation mechanism highlight diversification of the microsporidian ribosome. PLOS Biology 18, e3000958, https://doi.org/10.1371/journal.pbio.3000958 (2020).
Minato, T. et al. Non-conventional octameric structure of c-phycocyanin. Communications Biology 4, https://doi.org/10.1038/s42003-021-02767-x (2021).
Kato, T. et al. Structural insights into inhibitory mechanism of human excitatory amino acid transporter eaat2. Nature Communications 13, https://doi.org/10.1038/s41467-022-32442-6 (2022).
Hagino, T. et al. Cryo-em structures of thylakoid-located voltage-dependent chloride channel vccn1. Nature Communications 13, https://doi.org/10.1038/s41467-022-30292-w (2022).
Xiang, Y. et al. Superimmunity by pan-sarbecovirus nanobodies. Cell Reports 39, 111004, https://doi.org/10.1016/j.celrep.2022.111004 (2022).
Yamaguchi, S. et al. Structure of the dicer-2–r2d2 heterodimer bound to a small rna duplex. Nature 607, 393–398, https://doi.org/10.1038/s41586-022-04790-2 (2022).
B, B. et al. Apoferritin structure at 1.46 angstrom resolution by cryoarm300ii equipped with apollo https://doi.org/10.6019/EMPIAR-11101 (2022).
Nakagawa, R. et al. Structure and engineering of the minimal type vi crispr-cas13bt3. Molecular Cell 82, 3178–3192.e5, https://doi.org/10.1016/j.molcel.2022.08.001 (2022).
Jespersen, N. et al. Structure of the reduced microsporidian proteasome bound by pi31-like peptides in dormant spores. Nature Communications 13, https://doi.org/10.1038/s41467-022-34691-x (2022).
Suno, R. et al. Structural insights into the g protein selectivity revealed by the human ep3-gi signaling complex. Cell Reports 40, 111323, https://doi.org/10.1016/j.celrep.2022.111323 (2022).
Mori, T. et al. C-glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of c-c bond-cleaving enzymes. Nature Communications 12, https://doi.org/10.1038/s41467-021-26585-1 (2021).
Altomare, C. G. et al. Structure of a vaccine-induced, germline-encoded human antibody defines a neutralizing epitope on the sars-cov-2 spike n-terminal domain. mBio 13, https://doi.org/10.1128/mbio.03580-21 (2022).
Wang, L., Wu, D., Robinson, C. V., Wu, H. & Fu, T.-M. Structures of a complete human v-atpase reveal mechanisms of its assembly. Molecular Cell 80, 501–511.e3, https://doi.org/10.1016/j.molcel.2020.09.029 (2020).
Robertson, M. J. et al. Structure determination of inactive-state gpcrs with a universal nanobody https://doi.org/10.1101/2021.11.02.466983 (2021).
IB, S., J, P., JS, F. & DJ, L. E. coli 50s ribosome bound to d-linker solithromycin conjugate https://doi.org/10.6019/EMPIAR-11152 (2022).
Zhang, Z. et al. Structure of sars-cov-2 membrane protein essential for virus assembly. Nature Communications 13, https://doi.org/10.1038/s41467-022-32019-3 (2022).
Ikegaya, M. et al. Structural basis of the strict specificity of a bacterial gh31 α-1,3-glucosidase for nigerooligosaccharides. Journal of Biological Chemistry 298, 101827, https://doi.org/10.1016/j.jbc.2022.101827 (2022).
Ni, D. et al. Cryo-em structures and binding of mouse and human ace2 to sars-cov-2 variants of concern indicate that mutations enabling immune escape could expand host range. PLOS Pathogens 19, e1011206, https://doi.org/10.1371/journal.ppat.1011206 (2023).
Park, J.-H. et al. Structural insights into the hbv receptor and bile acid transporter ntcp. Nature 606, 1027–1031, https://doi.org/10.1038/s41586-022-04857-0 (2022).
Wilson, L. F. L. et al. The structure of extl3 helps to explain the different roles of bi-domain exostosins in heparan sulfate synthesis. Nature Communications 13, https://doi.org/10.1038/s41467-022-31048-2 (2022).
Muccini, A. J., Gustafson, M. A. & Fromme, J. C. Structural basis for activation of arf1 at the golgi complex. Cell Reports 40, 111282, https://doi.org/10.1016/j.celrep.2022.111282 (2022).
Spellmon, N. et al. Molecular basis for polysaccharide recognition and modulated atp hydrolysis by the o antigen abc transporter. Nature Communications 13, https://doi.org/10.1038/s41467-022-32597-2 (2022).
Podgorski, J. M. et al. A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability. Structure 31, 282–294.e5, https://doi.org/10.1016/j.str.2022.12.012 (2023).
Futamata, H. et al. Cryo-em structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility. Nature Communications 13, https://doi.org/10.1038/s41467-022-34017-x (2022).
Juyoux, P. et al. Architecture of the mkk6-p38α complex defines the basis of mapk specificity and activation. Science 381, 1217–1225, https://doi.org/10.1126/science.add7859 (2023).
Kishikawa, J.-i. et al. Cryo-em structures of na+-pumping nadh-ubiquinone oxidoreductase from vibrio cholerae. Nature Communications 13, https://doi.org/10.1038/s41467-022-31718-1 (2022).
Bacic, L. et al. Asymmetric nucleosome parylation at dna breaks mediates directional nucleosome sliding by alc1. Nature Communications 15, https://doi.org/10.1038/s41467-024-45237-8 (2024).
Bongiovanni, G., Harder, O. F., Drabbels, M. & Lorenz, U. J. Microsecond melting and revitrification of cryo samples with a correlative light-electron microscopy approach. Frontiers in Molecular Biosciences 9, https://doi.org/10.3389/fmolb.2022.1044509 (2022).
Zhao, H., Lee, J. & Chen, J. The hemolysin a secretion system is a multi-engine pump containing three abc transporters. Cell 185, 3329–3340.e13, https://doi.org/10.1016/j.cell.2022.07.017 (2022).
Jouravleva, K. et al. Structural basis of microrna biogenesis by dicer-1 and its partner protein loqs-pb. Molecular Cell 82, 4049–4063.e6, https://doi.org/10.1016/j.molcel.2022.09.002 (2022).
Maldonado, M., Fan, Z., Abe, K. M. & Letts, J. A. Plant-specific features of respiratory supercomplex i + iii2 from vigna radiata. Nature Plants 9, 157–168, https://doi.org/10.1038/s41477-022-01306-8 (2022).
Silberberg, J. M. et al. Inhibited kdpfabc transitions into an e1 off-cycle state. eLife 11, https://doi.org/10.7554/elife.80988 (2022).
Kawamoto, A. et al. Cryo-em structures of the translocational binary toxin complex cdta-bound cdtb-pore from clostridioides difficile. Nature Communications 13, https://doi.org/10.1038/s41467-022-33888-4 (2022).
Michalczyk, E. et al. Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin. Nature Catalysis 6, 52–67, https://doi.org/10.1038/s41929-022-00904-1 (2023).
E, M. & D, G. Escherichia coli gyrase holocomplex with 217 bp phage mu sgs dna and albi-1 stabilised by adpnp https://doi.org/10.6019/EMPIAR-11245 (2022).
E, M. & D, G. Escherichia coli gyrase holocomplex with 217 bp phage mu sgs dna and albi-2 stabilised by adpnp https://doi.org/10.6019/EMPIAR-11246 (2022).
Feathers, J. R., Richael, E. K., Simanek, K. A., Fromme, J. C. & Paczkowski, J. E. Structure of the rhlr-pqse complex from pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation. Structure 30, 1626–1636.e4, https://doi.org/10.1016/j.str.2022.10.008 (2022).
Young, V. C. et al. Structure and function of h+/k+ pump mutants reveal na+/k+ pump mechanisms. Nature Communications 13, https://doi.org/10.1038/s41467-022-32793-0 (2022).
C.-C., S. Cryo-em spa dataset of a native lysate fraction from human liver microsomes (fraction #1) https://doi.org/10.6019/EMPIAR-11249 (2022).
C.-C., S. Cryo-em spa dataset of a native lysate fraction from human liver microsomes (fraction #2) https://doi.org/10.6019/EMPIAR-11250 (2022).
Morgan, C. E., Zhang, Z., Miyagi, M., Golczak, M. & Yu, E. W. Toward structural-omics of the bovine retinal pigment epithelium. Cell Reports 41, 111876, https://doi.org/10.1016/j.celrep.2022.111876 (2022).
Laverty, D. et al. Cryo-em structure of the human α1β3 γ2 gabaa receptor in a lipid bilayer. Nature 565, 516–520, https://doi.org/10.1038/s41586-018-0833-4 (2019).
Masiulis, S. et al. Gabaa receptor signalling mechanisms revealed by structural pharmacology. Nature 565, 454–459, https://doi.org/10.1038/s41586-018-0832-5 (2019).
Peng, R. et al. Characterizing the resolution and throughput of the apollo direct electron detector. Journal of Structural Biology: X 7, 100080, https://doi.org/10.1016/j.yjsbx.2022.100080 (2023).
Yamada, T. et al. Cryo-em structures reveal translocational unfolding in the clostridial binary iota toxin complex. Nature Structural & Molecular Biology 27, 288–296, https://doi.org/10.1038/s41594-020-0388-6 (2020).
Chou, T.-H. et al. Structural insights into binding of therapeutic channel blockers in nmda receptors. Nature Structural & Molecular Biology 29, 507–518, https://doi.org/10.1038/s41594-022-00772-0 (2022).
Padavannil, A., Murari, A., Rhooms, S.-K., Owusu-Ansah, E. & Letts, J. A. Resting mitochondrial complex i from drosophila melanogaster adopts a helix-locked state. eLife 12, https://doi.org/10.7554/elife.84415 (2023).
H, V. 1.42 angstrom apoferritin structure determined using g1 titan krios s-feg operated at 300kv, zero loss imaging using gatan bioquantum energy filter operated at 10ev slit width and imaged using k2 camera. https://doi.org/10.6019/EMPIAR-11281 (2022).
Kern, D. M. et al. Structural basis for assembly and lipid-mediated gating of lrrc8a:c volume-regulated anion channels https://doi.org/10.1101/2022.07.31.502239 (2022).
JM, P. & SJ, W. Mycobacterium phage che8 mutant capsid (gene 110 deletion) https://doi.org/10.6019/EMPIAR-11285 (2022).
Yoshikawa, T. et al. Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis. Chemical Communications 58, 6478–6481, https://doi.org/10.1039/d2cc01541b (2022).
Jain, S. et al. Modulation of translational decoding by m6a modification of mrna. Nature Communications 14, https://doi.org/10.1038/s41467-023-40422-7 (2023).
Semchonok, D. A., Kyrilis, F. L., Hamdi, F. & Kastritis, P. L. Cryo-em of a heterogeneous cellular fraction elucidates multiple endogenous protein complexes from a thermophilic eukaryote. SSRN Electronic Journal https://doi.org/10.2139/ssrn.4211492 (2022).
Rennie, M. L., Arkinson, C., Chaugule, V. K. & Walden, H. Cryo-em reveals a mechanism of usp1 inhibition through a cryptic binding site. Science Advances 8, https://doi.org/10.1126/sciadv.abq6353 (2022).
Rennie, M. L., Arkinson, C., Chaugule, V. K., Toth, R. & Walden, H. Structural basis of fancd2 deubiquitination by usp1-uaf1. Nature Structural & Molecular Biology 28, 356–364, https://doi.org/10.1038/s41594-021-00576-8 (2021).
Thangaratnarajah, C. et al. Expulsion mechanism of the substrate-translocating subunit in ecf transporters. Nature Communications 14, https://doi.org/10.1038/s41467-023-40266-1 (2023).
Harper, N. J., Burnside, C. & Klinge, S. Principles of mitoribosomal small subunit assembly in eukaryotes. Nature 614, 175–181, https://doi.org/10.1038/s41586-022-05621-0 (2022).
Chung, K. et al. Structures of a mobile intron retroelement poised to attack its structured dna target. Science 378, 627–634, https://doi.org/10.1126/science.abq2844 (2022).
Hillen, H. S.Cryo-EM for Structure Determination of Mitochondrial Ribosome Samples, 89–100 https://doi.org/10.1007/978-1-0716-3171-3_6 (Springer US, 2023).
Hu, S. et al. Cryoelectron microscopic structure of the nucleoprotein–rna complex of the european filovirus, lloviu virus. PNAS Nexus 2, https://doi.org/10.1093/pnasnexus/pgad120 (2023).
Wang, W. & Pyle, A. M. The rig-i receptor adopts two different conformations for distinguishing host from viral rna ligands. Molecular Cell 82, 4131–4144.e6, https://doi.org/10.1016/j.molcel.2022.09.029 (2022).
Linares, R. et al. Structural basis of bacteriophage t5 infection trigger and e. coli cell wall perforation https://doi.org/10.1101/2022.09.20.507954 (2022).
Zhu, J. et al. Structural and mechanistic basis for recognition of alternative trna precursor substrates by bacterial ribonuclease p. Nature Communications 13, https://doi.org/10.1038/s41467-022-32843-7 (2022).
Liu, L. et al. Antibodies targeting a quaternary site on sars-cov-2 spike glycoprotein prevent viral receptor engagement by conformational locking. Immunity 56, 2442–2455.e8, https://doi.org/10.1016/j.immuni.2023.09.003 (2023).
Sutton, M. S. et al. Vaccine elicitation and structural basis for antibody protection against alphaviruses. Cell 186, 2672–2689.e25, https://doi.org/10.1016/j.cell.2023.05.019 (2023).
Ito, F. et al. Structural basis for hiv-1 antagonism of host apobec3g via cullin e3 ligase. Science Advances 9, https://doi.org/10.1126/sciadv.ade3168 (2023).
Chen, J. et al. Structure of an endogenous mycobacterial mce lipid transporter https://doi.org/10.21203/rs.3.rs-2412186/v1 (2023).
Bongiovanni, G., Harder, O. F., Voss, J. M., Drabbels, M. & Lorenz, U. J. Near-atomic resolution reconstructions from in situ revitrified cryo samples. Acta Crystallographica Section D Structural Biology 79, 473–478, https://doi.org/10.1107/s2059798323003431 (2023).
Wilson, S. C. et al. Organizing structural principles of the il-17 ligand–receptor axis. Nature 609, 622–629, https://doi.org/10.1038/s41586-022-05116-y (2022).
Sarewicz, M. et al. High-resolution cryo-em structures of plant cytochrome b6f at work. Science Advances 9, https://doi.org/10.1126/sciadv.add9688 (2023).
Caldwell, B. J. et al. Structure of a rect/redβ family recombinase in complex with a duplex intermediate of dna annealing. Nature Communications 13, https://doi.org/10.1038/s41467-022-35572-z (2022).
Ghanbarpour, A., Fei, X., Baker, T. A., Davis, J. H. & Sauer, R. T. The sspb adaptor drives structural changes in the aaa+clpxp protease during ssra-tagged substrate delivery https://doi.org/10.1101/2022.11.06.515074 (2022).
Akasaka, H. et al. Structure of the active gi-coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist. Nature Communications 13, https://doi.org/10.1038/s41467-022-33121-2 (2022).
Nureki, I. et al. Cryo-em structures of the β3 adrenergic receptor bound to solabegron and isoproterenol. Biochemical and Biophysical Research Communications 611, 158–164, https://doi.org/10.1016/j.bbrc.2022.04.065 (2022).
Nagiri, C. et al. Cryo-em structure of the β3-adrenergic receptor reveals the molecular basis of subtype selectivity. Molecular Cell 81, 3205–3215.e5, https://doi.org/10.1016/j.molcel.2021.06.024 (2021).
Weckener, M. et al. The lipid linked oligosaccharide polymerase wzy and its regulating co-polymerase, wzz, from enterobacterial common antigen biosynthesis form a complex. Open Biology 13, https://doi.org/10.1098/rsob.220373 (2023).
Yin, Z., Agip, A.-N. A., Bridges, H. R. & Hirst, J. Structural insights into respiratory complex i deficiency and assembly from the mitochondrial disease-related ndufs4-/- mouse. The EMBO Journal 43, 225–249, https://doi.org/10.1038/s44318-023-00001-4 (2024).
He, Q. et al. Structures of the human cst-polα–primase complex bound to telomere templates. Nature 608, 826–832, https://doi.org/10.1038/s41586-022-05040-1 (2022).
He, Q. et al. Structures of human primosome elongation complexes. Nature Structural & Molecular Biology 30, 579–583, https://doi.org/10.1038/s41594-023-00971-3 (2023).
Tucker, K., Sridharan, S., Adesnik, H. & Brohawn, S. G. Cryo-em structures of the channelrhodopsin chrmine in lipid nanodiscs. Nature Communications 13, https://doi.org/10.1038/s41467-022-32441-7 (2022).
Domanska, A. et al. Structural studies reveal that endosomal cations promote formation of infectious coxsackievirus a9 a-particles, facilitating rna and vp4 release. Journal of Virology 96, https://doi.org/10.1128/jvi.01367-22 (2022).
McLaren, M. et al. Cryoem reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state. Nature Microbiology 8, 1834–1845, https://doi.org/10.1038/s41564-023-01469-w (2023).
Lemonidis, K. et al. Structural and biochemical basis of interdependent fanci-fancd2 ubiquitination. The EMBO Journal 42, https://doi.org/10.15252/embj.2022111898 (2022).
Reimer, J. M., DeSantis, M. E., Reck-Peterson, S. L. & Leschziner, A. E. Structures of human dynein in complex with the lissencephaly 1 protein, lis1. eLife 12, https://doi.org/10.7554/elife.84302 (2023).
Chou, T.-H., Kang, H., Simorowski, N., Traynelis, S. F. & Furukawa, H. Structural insights into assembly and function of glun1-2c, glun1-2a-2c, and glun1-2d nmdars. Molecular Cell 82, 4548–4563.e4, https://doi.org/10.1016/j.molcel.2022.10.008 (2022).
Fromm, S. A. et al. The translating bacterial ribosome at 1.55 Å resolution by open access cryo-em https://doi.org/10.1101/2022.08.30.505838 (2022).
ZA, S., R, P., A, V. B. & S, K. The noc1-noc2 rnp - a co-transcriptional large ribosomal assembly intermediate https://doi.org/10.6019/EMPIAR-11379 (2023).
P, M. & G, P. Enp1-tap associated immature ribosomal particles from s. cerevisiae https://doi.org/10.6019/EMPIAR-11387 (2022).
Koller, T. O. et al. Structural basis for translation inhibition by the glycosylated drosocin peptide. Nature Chemical Biology 19, 1072–1081, https://doi.org/10.1038/s41589-023-01293-7 (2023).
P, M. & G, P. Enp1-tap associated immature ribosomal particles from s. cerevisiae depleted of rps21 https://doi.org/10.6019/EMPIAR-11389 (2023).
P, M. & G, P. Slx9-tap associated immature ribosomal particles from s. cerevisiae depleted of rps21 https://doi.org/10.6019/EMPIAR-11390 (2023).
Miotto, M. C. et al. Structural analyses of human ryanodine receptor type 2 channels reveal the mechanisms for sudden cardiac death and treatment. Science Advances 8, https://doi.org/10.1126/sciadv.abo1272 (2022).
Tao, H. et al. Discovery of non-squalene triterpenes. Nature 606, 414–419, https://doi.org/10.1038/s41586-022-04773-3 (2022).
S, S. et al. Unaligned cryo-em micrographs of human shmt1 in complex with rna https://doi.org/10.6019/EMPIAR-11413 (2023).
Velilla, J. A. et al. Structural basis of colibactin activation by the clbp peptidase. Nature Chemical Biology 19, 151–158, https://doi.org/10.1038/s41589-022-01142-z (2022).
Ito, F., Alvarez-Cabrera, A. L., Kim, K., Zhou, Z. H. & Chen, X. S. Structural basis of hiv-1 vif-mediated e3 ligase targeting of host apobec3h. Nature Communications 14, https://doi.org/10.1038/s41467-023-40955-x (2023).
Chio, U. S. et al. Cryo-em structure of the human sirtuin 6–nucleosome complex. Science Advances 9, https://doi.org/10.1126/sciadv.adf7586 (2023).
Ehrmann, J. F. et al. Structural basis of how the birc6/smac complex regulates apoptosis and autophagy https://doi.org/10.1101/2022.08.30.505823 (2022).
Barros-Álvarez, X. et al. The tethered peptide activation mechanism of adhesion gpcrs. Nature 604, 757–762, https://doi.org/10.1038/s41586-022-04575-7 (2022).
Cao, C. et al. Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic lsd. Neuron 110, 3154–3167.e7, https://doi.org/10.1016/j.neuron.2022.08.006 (2022).
Kaplan, A. L. et al. Bespoke library docking for 5-ht2a receptor agonists with antidepressant activity. Nature 610, 582–591, https://doi.org/10.1038/s41586-022-05258-z (2022).
He, C. et al. Cd19 car antigen engagement mechanisms and affinity tuning. Science Immunology 8, https://doi.org/10.1126/sciimmunol.adf1426 (2023).
Deák, G. et al. Histone divergence in trypanosomes results in unique alterations to nucleosome structure. Nucleic Acids Research 51, 7882–7899, https://doi.org/10.1093/nar/gkad577 (2023).
S. Cannon, K., Sarsam, R. D., Tedamrongwanish, T., Zhang, K. & Baker, R. W. Lipid nanodiscs as a template for high-resolution cryo-em structures of peripheral membrane proteins. Journal of Structural Biology 215, 107989, https://doi.org/10.1016/j.jsb.2023.107989 (2023).
Wilkinson, M. E., Frangieh, C. J., Macrae, R. K. & Zhang, F. Structure of the r2 non-ltr retrotransposon initiating target-primed reverse transcription. Science 380, 301–308, https://doi.org/10.1126/science.adg7883 (2023).
TI, K. & AJ, J. Cryo-em structure of the human gbp1 dimer bound to gdp-alf3 https://doi.org/10.6019/EMPIAR-11459 (2023).
G, B., OF, H., JM, V., M, D. & UJ, L. Mouse heavy chain apoferritin in vitreous ice after laser-melting and revitrification https://doi.org/10.6019/EMPIAR-11460 (2022).
Harder, O. F., Barrass, S. V., Drabbels, M. & Lorenz, U. J. Fast viral dynamics revealed by microsecond time-resolved cryo-em. Nature Communications 14, https://doi.org/10.1038/s41467-023-41444-x (2023).
Strecker, J. et al. Rna-activated protein cleavage with a crispr-associated endopeptidase. Science 378, 874–881, https://doi.org/10.1126/science.add7450 (2022).
X, H. et al. Cryoem structure of aspergillus nidulans utp-glucose-1-phosphate uridylyltransferase https://doi.org/10.6019/EMPIAR-11471 (2022).
Krishna Kumar, K. et al. Structural basis for activation of cb1 by an endocannabinoid analog. Nature Communications 14, https://doi.org/10.1038/s41467-023-37864-4 (2023).
Yang, K. et al. Nanomolar inhibition of sars-cov-2 infection by an unmodified peptide targeting the prehairpin intermediate of the spike protein. Proceedings of the National Academy of Sciences 119, https://doi.org/10.1073/pnas.2210990119 (2022).
Yang, K. et al. Structure-based design of a sars-cov-2 omicron-specific inhibitor. Proceedings of the National Academy of Sciences 120, https://doi.org/10.1073/pnas.2300360120 (2023).
Kilkenny, M. L. et al. Structural basis for the interaction of sars-cov-2 virulence factor nsp1 with dna polymerase α–primase. Protein Science 31, 333–344, https://doi.org/10.1002/pro.4220 (2021).
Torino, S., Dhurandhar, M., Stroobants, A., Claessens, R. & Efremov, R. G. Time-resolved cryo-em using a combination of droplet microfluidics with on-demand jetting. Nature Methods 20, 1400–1408, https://doi.org/10.1038/s41592-023-01967-z (2023).
El Mazouni, D. & Gros, P. Cryo-em structures of peripherin-2 and rom1 suggest multiple roles in photoreceptor membrane morphogenesis. Science Advances 8, https://doi.org/10.1126/sciadv.add3677 (2022).
Caveney, N. A., Glassman, C. R., Jude, K. M., Tsutsumi, N. & Garcia, K. C. Structure of the il-27 quaternary receptor signaling complex. eLife 11, https://doi.org/10.7554/elife.78463 (2022).
H, Q. Structure of human choline/ethanolamine phosphotransferase https://doi.org/10.6019/EMPIAR-11492 (2023).
Wang, W., Götte, B., Guo, R. & Pyle, A. M. The e3 ligase riplet promotes rig-i signaling independent of rig-i oligomerization. Nature Communications 14, https://doi.org/10.1038/s41467-023-42982-0 (2023).
W, W. & AM, P. Single particle cryo-em structure of 14-aa-gs-rig-i in complex with p3slr30 https://doi.org/10.6019/EMPIAR-11495 (2023).
W, W. & AM, P. Single particle cryo-em structure of rig-i in complex with p3slr14 https://doi.org/10.6019/EMPIAR-11496 (2023).
JM, P. Mycobacterium phage patience https://doi.org/10.6019/EMPIAR-11498 (2023).
JM, P. Mycobacterium phage adjutor https://doi.org/10.6019/EMPIAR-11499 (2023).
Kang, J. Y. et al. An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options. Proceedings of the National Academy of Sciences 120, https://doi.org/10.1073/pnas.2215945120 (2023).
Wang, P. et al. A monoclonal antibody that neutralizes sars-cov-2 variants, sars-cov, and other sarbecoviruses https://doi.org/10.1080/22221751.2021.2011623 (2021).
Grba, D. N., Chung, I., Bridges, H. R., Agip, A.-N. A. & Hirst, J. Investigation of hydrated channels and proton pathways in a high-resolution cryo-em structure of mammalian complex i. Science Advances 9, https://doi.org/10.1126/sciadv.adi1359 (2023).
Maki-Yonekura, S., Kawakami, K., Takaba, K., Hamaguchi, T. & Yonekura, K. Measurement of charges and chemical bonding in a cryo-em structure. Communications Chemistry 6, 98, https://doi.org/10.1038/s42004-023-00900-x (2023).
Durieux Trouilleton, Q., Barata-García, S., Arragain, B., Reguera, J. & Malet, H. Structures of active hantaan virus polymerase uncover the mechanisms of hantaviridae genome replication. Nature Communications 14, 2954, https://doi.org/10.1038/s41467-023-38555-w (2023).
Kavčič, L. et al. From structural polymorphism to structural metamorphosis of the coat protein of flexuous filamentous potato virus y. Communications Chemistry 7, 14, https://doi.org/10.1038/s42004-024-01100-x (2024).
Sverzhinsky, A., Tomkinson, A. E. & Pascal, J. M. Cryo-em structures and biochemical insights into heterotrimeric pcna regulation of dna ligase. Structure 30, 371–385.e5, https://doi.org/10.1016/j.str.2021.11.002 (2022).
Tajima, S. et al. Structural basis for ion selectivity in potassium-selective channelrhodopsins. Cell 186, 4325–4344.e26, https://doi.org/10.1016/j.cell.2023.08.009 (2023).
Ni, T. et al. Intrinsically disordered csos2 acts as a general molecular thread for α-carboxysome shell assembly. Nature Communications 14, 5512, https://doi.org/10.1038/s41467-023-41211-y (2023).
Caveney, N. A., Tsutsumi, N. & Garcia, K. C. Structural insight into guanylyl cyclase receptor hijacking of the kinase–hsp90 regulatory mechanism https://doi.org/10.1101/2023.02.14.528495 (2023).
Highland, C. M., Tan, A., Ricaña, C. L., Briggs, J. A. G. & Dick, R. A. Structural insights into hiv-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-em. Proceedings of the National Academy of Sciences 120, https://doi.org/10.1073/pnas.2220545120 (2023).
Rybak, M. Y. & Gagnon, M. G. Structures of the ribosome bound to ef-tu–isoleucine trna elucidate the mechanism of aug avoidance. Nature Structural & Molecular Biology 31, 810–816, https://doi.org/10.1038/s41594-024-01236-3 (2024).
Abeywansha, T. et al. The structural basis of trna recognition by arginyl-trna-protein transferase. Nature Communications 14, https://doi.org/10.1038/s41467-023-38004-8 (2023).
Kamegawa, A. et al. Structural analysis of the water channel aqp2 by single-particle cryo-em. Journal of Structural Biology 215, 107984, https://doi.org/10.1016/j.jsb.2023.107984 (2023).
Absmeier, E. et al. Specific recognition and ubiquitination of slow-moving ribosomes by human ccr4-not https://doi.org/10.1101/2022.07.24.501325 (2022).
PK, S. & TM, I. Structural basis for directional rotation of the salmonella flagellum https://doi.org/10.6019/EMPIAR-11597 (2023).
HW, Q. Raw images of human cept1 complexed with cdp-choline https://doi.org/10.6019/EMPIAR-11600 (2023).
T, O., H, Y. & M, K. Porcine uroplakin complex https://doi.org/10.6019/EMPIAR-11601 (2023).
Li, J. et al. Alternative splicing controls teneurin-latrophilin interaction and synapse specificity by a shape-shifting mechanism. Nature Communications 11, https://doi.org/10.1038/s41467-020-16029-7 (2020).
Zhang, J., Maksaev, G. & Yuan, P. Open structure and gating of the arabidopsis mechanosensitive ion channel msl10. Nature Communications 14, https://doi.org/10.1038/s41467-023-42117-5 (2023).
G, G. Single-particle cryo-em dataset of an asymmetric nucleosome (2023 (10-n-0). https://doi.org/10.6019/EMPIAR-11618.
H, S. & Y, F. The j-k rna fragment of encephalomyocarditis virus ires in complex with eif4g-heat1 and eif4a https://doi.org/10.6019/EMPIAR-11624 (2023).
Bavi, N. et al. The conformational cycle of prestin underlies outer-hair cell electromotility. Nature 600, 553–558, https://doi.org/10.1038/s41586-021-04152-4 (2021).
Kalienkova, V., Peter, M. F., Rheinberger, J. & Paulino, C. Structures of a sperm-specific solute carrier gated by voltage and camp. Nature 623, 202–209, https://doi.org/10.1038/s41586-023-06629-w (2023).
Kalienkova, V., Dandamudi, M., Paulino, C. & Lynagh, T. Structural basis for excitatory neuropeptide signaling. Nature Structural & Molecular Biology 31, 717–726, https://doi.org/10.1038/s41594-023-01198-y (2024).
V, K., MF, P., J, R. & C, P. camp-bound spslc9c1 in lipid nanodiscs https://doi.org/10.6019/EMPIAR-11635 (2023).
Adachi, T. et al. Experimental and theoretical insights into bienzymatic cascade for mediatorless bioelectrochemical ethanol oxidation with alcohol and aldehyde dehydrogenases. ACS Catalysis 13, 7955–7965, https://doi.org/10.1021/acscatal.3c01962 (2023).
Wang, T. et al. Fenofibrate recognition and gq protein coupling mechanisms of the human cannabinoid receptor cb1. Advanced Science 11, https://doi.org/10.1002/advs.202306311 (2024).
Sauer, P. V. et al. Structural and quantum chemical basis for ocp-mediated quenching of phycobilisomes. Science Advances 10, https://doi.org/10.1126/sciadv.adk7535 (2024).
Watanabe, S. et al. Structure of full-length ergic-53 in complex with mcfd2 for cargo transport. Nature Communications 15, https://doi.org/10.1038/s41467-024-46747-1 (2024).
Agip, A.-N. A. et al. Cryo-em structures of complex i from mouse heart mitochondria in two biochemically defined states. Nature Structural & Molecular Biology 25, 548–556, https://doi.org/10.1038/s41594-018-0073-1 (2018).
Blaza, J. N., Vinothkumar, K. R. & Hirst, J. Structure of the deactive state of mammalian respiratory complex i. Structure 26, 312–319.e3, https://doi.org/10.1016/j.str.2017.12.014 (2018).
Agip, A.-N. A., Chung, I., Sanchez-Martinez, A., Whitworth, A. J. & Hirst, J. Cryo-em structures of mitochondrial respiratory complex i from drosophila melanogaster. eLife 12, https://doi.org/10.7554/elife.84424 (2023).
Chung, I. et al. Cork-in-bottle mechanism of inhibitor binding to mammalian complex i. Science Advances 7, https://doi.org/10.1126/sciadv.abg4000 (2021).
Santiago-Frangos, A. et al. Structure reveals why genome folding is necessary for site-specific integration of foreign dna into crispr arrays. Nature Structural & Molecular Biology 30, 1675–1685, https://doi.org/10.1038/s41594-023-01097-2 (2023).
Bodrug, T. et al. Time-resolved cryo-em (tr-em) analysis of substrate polyubiquitination by the ring e3 anaphase-promoting complex/cyclosome (apc/c). Nature Structural & Molecular Biology 30, 1663–1674, https://doi.org/10.1038/s41594-023-01105-5 (2023).
Rüttermann, M. et al. Structure of the peroxisomal pex1/pex6 atpase complex bound to a substrate. Nature Communications 14, https://doi.org/10.1038/s41467-023-41640-9 (2023).
Agip, A.-N. A., Blaza, J. N., Fedor, J. G. & Hirst, J. Mammalian respiratory complex i through the lens of cryo-em. Annual Review of Biophysics 48, 165–184, https://doi.org/10.1146/annurev-biophys-052118-115704 (2019).
Dolce, L. G. et al. Structural basis for sequence-independent substrate selection by eukaryotic wobble base trna deaminase adat2/3. Nature Communications 13, https://doi.org/10.1038/s41467-022-34441-z (2022).
Dolce, L. G., Nesterenko, Y., Walther, L., Weis, F. & Kowalinski, E. Structural basis for guide rna selection by the resc1–resc2 complex. Nucleic Acids Research 51, 4602–4612, https://doi.org/10.1093/nar/gkad217 (2023).
Carman, P. J., Barrie, K. R., Rebowski, G. & Dominguez, R. Structures of the free and capped ends of the actin filament. Science 380, 1287–1292, https://doi.org/10.1126/science.adg6812 (2023).
Nygaard, R. et al. Structural basis of peptidoglycan synthesis by e. coli roda-pbp2 complex. Nature Communications 14, https://doi.org/10.1038/s41467-023-40483-8 (2023).
Suzuki, S. et al. Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding. Nature Communications 14, https://doi.org/10.1038/s41467-023-41650-7 (2023).
Ghanbarpour, A. et al. A closed translocation channel in the substrate-free aaa+ clpxp protease diminishes rogue degradation. Nature Communications 14, https://doi.org/10.1038/s41467-023-43145-x (2023).
Paidimuddala, B. et al. Mechanism of naip-nlrc4 inflammasome activation revealed by cryo-em structure of unliganded naip5. Nature Structural & Molecular Biology 30, 159–166, https://doi.org/10.1038/s41594-022-00889-2 (2023).
Xu, T.-H. et al. Structure of nucleosome-bound dna methyltransferases dnmt3a and dnmt3b. Nature 586, 151–155, https://doi.org/10.1038/s41586-020-2747-1 (2020).
Sijacki, T. et al. The dna-damage kinase atr activates the fancd2-fanci clamp by priming it for ubiquitination. Nature Structural & Molecular Biology 29, 881–890, https://doi.org/10.1038/s41594-022-00820-9 (2022).
Seifert-Davila, W. et al. Structural insights into human tfiiic promoter recognition. Science Advances 9, https://doi.org/10.1126/sciadv.adh2019 (2023).
Zhao, F., Hicks, C. W. & Wolberger, C. Mechanism of histone h2b monoubiquitination by bre1. Nature Structural & Molecular Biology 30, 1623–1627, https://doi.org/10.1038/s41594-023-01137-x (2023).
Paidimuddala, B., Cao, J. & Zhang, L. Structural basis for flagellin-induced naip5 activation. Science Advances 9, https://doi.org/10.1126/sciadv.adi8539 (2023).
Yang, L. et al. High resolution cryo-em and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase. Nature Communications 14, https://doi.org/10.1038/s41467-023-40253-6 (2023).
Pacesa, M. et al. R-loop formation and conformational activation mechanisms of cas9. Nature 609, 191–196, https://doi.org/10.1038/s41586-022-05114-0 (2022).
Gambelli, L. et al. Structure of the two-component s-layer of the archaeon sulfolobus acidocaldarius. eLife 13, https://doi.org/10.7554/elife.84617 (2024).
Appleby, R., Joudeh, L., Cobbett, K. & Pellegrini, L. Structural basis for stabilisation of the rad51 nucleoprotein filament by brca2. Nature Communications 14, https://doi.org/10.1038/s41467-023-42830-1 (2023).
Cushing, V. I. et al. High-resolution cryo-em of the human cdk-activating kinase for structure-based drug design. Nature Communications 15, https://doi.org/10.1038/s41467-024-46375-9 (2024).
Kretsch, R. C. et al. Tertiary folds of the sl5 rna from the 5’ proximal region of sars-cov-2 and related coronaviruses. Proceedings of the National Academy of Sciences 121, https://doi.org/10.1073/pnas.2320493121 (2024).
Kato, K. et al. Rna-triggered protein cleavage and cell growth arrest by the type iii-e crispr nuclease-protease. Science 378, 882–889, https://doi.org/10.1126/science.add7347 (2022).
Kato, K. et al. Structure and engineering of the type iii-e crispr-cas7-11 effector complex. Cell 185, 2324–2337.e16, https://doi.org/10.1016/j.cell.2022.05.003 (2022).
Hsu, H.-C. et al. Structures revealing mechanisms of resistance and collateral sensitivity of plasmodium falciparum to proteasome inhibitors. Nature Communications 14, https://doi.org/10.1038/s41467-023-44077-2 (2023).
DAmico, K. A. et al. Structure of a membrane tethering complex incorporating multiple snares https://doi.org/10.1101/2023.01.30.526244 (2023).
Kretsch, R. C. et al. Tertiary folds of the sl5 rna from the 5’ proximal region of sars-cov-2 and related coronaviruses https://doi.org/10.1101/2023.11.22.567964 (2023).
Wang, F., Feng, X., He, Q., Li, H. & Li, H. The saccharomyces cerevisiae yta7 atpase hexamer contains a unique bromodomain tier that functions in nucleosome disassembly. Journal of Biological Chemistry 299, 102852, https://doi.org/10.1016/j.jbc.2022.102852 (2023).
Papasergi-Scott, M. M. et al. Time-resolved cryo-em of g-protein activation by a gpcr. Nature 629, 1182–1191, https://doi.org/10.1038/s41586-024-07153-1 (2024).
Cerutti, G. et al. Structural basis for accommodation of emerging b.1.351 and b.1.1.7 variants by two potent sars-cov-2 neutralizing antibodies. Structure 29, 655–663.e4, https://doi.org/10.1016/j.str.2021.05.014 (2021).
S.U., N., A.G., M. & I., M. Cryo-em structure of mouse heavy-chain apoferritin https://doi.org/10.6019/EMPIAR-11866 (2023).
Cerutti, G. et al. Potent sars-cov-2 neutralizing antibodies directed against spike n-terminal domain target a single supersite. Cell Host & Microbe 29, 819–833.e7, https://doi.org/10.1016/j.chom.2021.03.005 (2021).
Sanz Murillo, M. et al. Inhibition of parkinson’s disease–related lrrk2 by type i and type ii kinase inhibitors: Activity and structures. Science Advances 9, https://doi.org/10.1126/sciadv.adk6191 (2023).
Cerutti, G. et al. Cryo-em structure of the sars-cov-2 omicron spike. Cell Reports 38, 110428, https://doi.org/10.1016/j.celrep.2022.110428 (2022).
Wang, F. et al. Structure of the human ubr5 e3 ubiquitin ligase. Structure 31, 541–552.e4, https://doi.org/10.1016/j.str.2023.03.010 (2023).
Xing, C. et al. Cryo-em structure of the human cannabinoid receptor cb2-gi signaling complex. Cell 180, 645–654.e13, https://doi.org/10.1016/j.cell.2020.01.007 (2020).
Cerutti, G. et al. Neutralizing antibody 5-7 defines a distinct site of vulnerability in sars-cov-2 spike n-terminal domain. Cell Reports 37, 109928, https://doi.org/10.1016/j.celrep.2021.109928 (2021).
Dendooven, T. et al. Cryo-em structure of the complete inner kinetochore of the budding yeast point centromere. Science Advances 9, https://doi.org/10.1126/sciadv.adg7480 (2023).
PK, S. & TM, I. Structural basis for directional rotation of the salmonella flagellum https://doi.org/10.6019/EMPIAR-11891 (2024).
TM, M. et al. cryo-em structure of nucleotide-free rmrp2 https://doi.org/10.6019/EMPIAR-11893 (2024).
TM, M. et al. cryo-em structure of rmrp2 in complex with probenecid https://doi.org/10.6019/EMPIAR-11894 (2024).
Blomgren, L. K. M. et al. Dynamic inter-domain transformations mediate the allosteric regulation of human 5, 10-methylenetetrahydrofolate reductase. Nature Communications 15, https://doi.org/10.1038/s41467-024-47174-y (2024).
Flynn, A. J., Antonyuk, S. V., Eady, R. R., Muench, S. P. & Hasnain, S. S. A 2.2 Å cryoem structure of a quinol-dependent no reductase shows close similarity to respiratory oxidases. Nature Communications 14, https://doi.org/10.1038/s41467-023-39140-x (2023).
Ghanim, G. E., Sekne, Z., Balch, S., van Roon, A.-M. M. & Nguyen, T. H. D. 2.7 Å cryo-em structure of human telomerase h/aca ribonucleoprotein. Nature Communications 15, https://doi.org/10.1038/s41467-024-45002-x (2024).
Vallet, S. D. et al. Functional and structural insights into human n-deacetylase/n-sulfotransferase activities. Proteoglycan Research 1, https://doi.org/10.1002/pgr2.8 (2023).
Uday, A. B., Mishra, R. K. & Hussain, T. Initiation factor 3 bound to the 30s ribosomal subunit in an initial step of translation. Proteins: Structure, Function, and Bioinformatics 93, 279–286, https://doi.org/10.1002/prot.26655 (2023).
Sun, C. et al. The 2.6 Å structure of a tulane virus variant with minor mutations leading to receptor change. Biomolecules 14, 119, https://doi.org/10.3390/biom14010119 (2024).
Kuklewicz, J. & Zimmer, J. Molecular insights into capsular polysaccharide secretion. Nature 628, 901–909, https://doi.org/10.1038/s41586-024-07248-9 (2024).
Fregoso, F. E. et al. Mechanism of synergistic activation of arp2/3 complex by cortactin and wasp-family proteins. Nature Communications 14, https://doi.org/10.1038/s41467-023-42229-y (2023).
Acknowledgements
This work was supported by the 2024 Shanghai Action for Science, Technology and Innovation Program of Natural Science Foundation of Shanghai grant nos. 24JS2820100 (Z.-J.L.), the HPC Platform of ShanghaiTech University. We thank EMPIAR and EMDB for providing cryo-EM image data and metadata, as well as the researchers who contributed data to these repositories. We thank the CryoSPARC team for their data processing platform, particularly the Micrograph Denoiser in CryoSPARC v4.5, which inspired our preprocessing pipeline. We thank L.-P. Sun for assistance with micrograph preprocessing.
Author information
Authors and Affiliations
Contributions
J.Y. conceived and conceptualized the research; Z.L., Y.P. and J.Y. provided guidance throughout the study; Q.C. and Y.P. designed the methodology and developed the code for data construction; Q.C. and Z.X. curated and visualized the data; Q.C. and H.D. wrote code to validate the data using DRACO; Q.C. drafted the manuscript, while J.Z., H.D., Y.S., Y.P., and J.Y. revised it; All authors contributed to the analysis of the data, discussed the results, and reviewed the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing interests: Jiakai Zhang is the co-founder of Cellverse, Co., Ltd., which develops cryo-EM related algorithms and may benefit from the publication of this research. Qihe Chen, Zhenyang Xu, Haizhao Dai, and Yingjun Shen were interns at Cellverse, Co., Ltd. during the course of this research. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, Q., Xu, Z., Dai, H. et al. A large-scale curated and filterable dataset for cryo-EM foundation model pre-training. Sci Data 12, 960 (2025). https://doi.org/10.1038/s41597-025-05179-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05179-2