Abstract
Accurate measurement of hallux valgus angle (HVA) and intermetatarsal angle (IMA) is essential for diagnosing hallux valgus and determining appropriate treatment strategies. Traditional manual measurement methods, while standardized, are time-consuming, labor-intensive, and subject to evaluator bias. Recent advancements in deep learning have been applied to hallux valgus angle estimation, but the development of effective algorithms requires large, well-annotated datasets. Existing X-ray datasets are typically limited to cropped foot regions images, and only one dataset containing very few samples is publicly available. To address these challenges, we introduce HVAngleEst, the first large-scale, open-access dataset specifically designed for hallux valgus angle estimation. HVAngleEst comprises 1,382 X-ray images from 1,150 patients and includes comprehensive annotations, such as foot localization, hallux valgus angles, and line segments for each phalanx. This dataset enables fully automated, end-to-end hallux valgus angle estimation, reducing manual labor and eliminating evaluator bias.
Similar content being viewed by others
Background & Summary
Hallux valgus is a common foot deformity in medical practice, often accompanied by significant functional impairment and foot pain. The prevalence of hallux valgus in females is 2.3 to 4.7 times that in males1,2. Accurate, reliable, and reproducible measurement of hallux valgus angle (HVA) and intermetatarsal angle (IMA) is essential for diagnosing hallux valgus and determining appropriate clinical surgical treatments3,4,5. Manual measurement methods for measuring HVA and IMA were standardized by the ad hoc committee of the American Orthopaedic Foot & Ankle Society (AOFAS)6,7. These standardized methods have been demonstrated to be reliable within a 5° margin. However, the potential for bias among different evaluators remains a challenge and manual methods are both time-consuming and labor-intensive.
Recent advancements in deep learning have revolutionized medical imaging tasks such as object detection and segmentation8,9,10,11,12,13. Several attempts have also been made to apply deep learning to hallux valgus angle estimation, and current methods typically approach the problem as a segmentation task followed by linear regression14,15,16 by using a cropped foot as input. This involves segmenting line segments representing the great toe, the first metatarsal, and the second metatarsal using segmentation networks. Subsequently, linear regression is applied to compute the HVA and IMA. Despite these innovations, training these models requires large, well-annotated datasets. This is particularly challenging in the medical field due to the sensitive nature of the data and the difficulty in making such datasets publicly available.
Existing studies on hallux valgus angle estimation are summarized in Table 1. Kwolek et al. pioneered the segmentation-based approach to HVA estimation but trained their model on only 30 images, focusing solely on HVA14. Xu et al. employed an Hourglass neural network trained on 230 X-ray images from 143 patients, with annotations that were not subjected to a rigorous annotation process16. Takeda et al. and Ma et al. conducted experiments on 1,798 and 2,000 images, respectively, to estimate HVA and IMA15. However, only 230 images from Xu et al. dataset are publicly accessible. Moreover, all these datasets are composed of cropped foot images, which limits their applicability to real-world clinical scenarios, particularly when an X-ray contains both feet or when the majority of the image is not occupied by a foot.
To overcome these challenges, we introduce HVAngleEst, the first large-scale, open-access annotated dataset designed for developing hallux valgus angle estimation algorithms. HVAngleEst comprises 1,382 X-ray images collected from the Foot and Ankle Surgery Department, Honghui Hospital of Xi’an Jiaotong University, China. It includes annotations of hallux valgus angles, feet localization and line segments representing the great toe, the first metatarsal, and the second metatarsal, on full X-ray images. This dataset enables fully automated, end-to-end hallux valgus angle estimation, minimizing manual effort.
Methods
This study was approved by the institutional ethics committee of Honghui Hospital of Xi’an Jiaotong University,the relevant case number is 2025-KY-017-01. As the dataset was collected retrospectively and all sensitive information related to patients was anonymized, the ethics committee waived the requirement for obtaining informed consent from patients. The entire data labeling pipeline is illustrated in Fig. 1.
The workflow for creating the HVAngleEst dataset: (1) collected X-ray images and converted to JPG by giving arbitrary names. (2) Removed the invalid images and crop the images that contain personal information on the edges. (3) Performed statistical analysis on the data obtained in the stage 2. (4) Four orthopedic doctors annotated the images from the stage 2, they manually annotated boxes and endpoints of line segment. Subsequently, the HVA and IMA were calculated from endpoints.
Data collection and cleanup
The HVAngleEst dataset includes 1,587 feet across 1,150 patients, captured in 1,382 X-ray images. The dataset comprises 130 male patients and 1020 female patients, with 825 left feet and 762 right feet. Patient ages range from 17 to 83 years (mean ± standard deviation: 51.4 ± 15.0 years). Of the 1,382 images, 1,332 DICOM images were generated using Siemens device, while 50 images were photographs of printed X-ray films taken with camera (Canon or Nikon). This inclusion of both high-quality DICOM images and mobile-captured film photographs reflects real-world clinical practice, where patients often present with images of printed X-rays from external institutions and the original DICOM files are unavailable. To minimize financial burdens, clinicians may choose to utilize these secondary images rather than request new acquisitions. By incorporating both high-quality DICOM images and mobile-captured film photographs into training dataset, we aim to replicate realistic clinical workflows, ensuring that the AI algorithm can robustly handle multi-source imaging data with varying quality and provenance. Current research demonstrates that properly designed multi-modal training not only fails to confuse the AI algorithm but instead can force it to learn more robust and generalizable features17. All photographs in our dataset underwent strict quality control, and we explicitly labeled the image source in the “source” column of datasets.csv, allowing users to filter by modality (DICOM vs. camera) if desired.
The dataset contains 620 images of the left foot, 558 images of the right foot, and 204 images showing both feet. A total of 366 feet were labeled with the property “truncated”, indicating partial occlusion or extension beyond the image boundary. DICOM images were anonymized by assigning arbitrary names and converting them to BMP format, with sensitive metadata removed. Subsequently, BMP and camera images were converted to JPG images, and files were renamed using a Python script and all images were manually reviewed to ensure privacy. During the manual inspection, personal information, such as names and birth dates, was found on the edges of 578 images. To ensure patient confidentiality, these images were manually cropped to remove any personal information.
Image annotation
The X-ray images were annotated using the LabelMe tool (https://github.com/wkentaro/labelme.git)18, and the annotation process was performed by four orthopedic doctors with varying levels of experience: Labeler A (3 years), Labeler B (5 years), Labeler C (over 10 years), and Labeler D (over 20 years).
For foot localization labeling, the IoU (Intersection over Union) between bounding boxes labeled by Labeler A and Labeler B was evaluated. If the IoU exceeded 0.95, Labeler B’s annotation was accepted. Otherwise, Labeler C reviewed the task. If the IoU between the bounding boxes annotated by Labeler C and those by either Labeler A or Labeler B exceeded 0.95, Labeler C’s annotation was considered final. Otherwise, the task was escalated to Labeler D, whose annotation was considered definitive. Bounding boxes were categorized as “left” or “right” foot, and the “truncated” property was assigned if the foot was either partially occluded or extended beyond the image boundary.
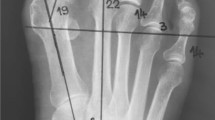
HVA, IMA and line segments were annotated in accordance with the standards set by AOFAS4,5, as shown in Fig. 2. First, two parallel lines (red dashed lines) were drawn for the great toe, the first metatarsal, and the second metatarsal. Second, the center lines of each phalanx (blue solid lines) were drawn through the midpoints of the two parallel lines. Third, the intersection points of these center lines with the ends of each phalanx were identified and served as the endpoints for the corresponding phalanx line segments Finally, the HVA and IMA were calculated using the labeled endpoints of each phalanx. If discrepancies between HVA and IMA annotations by Labelers A and B were less than 1°, Labeler B’s annotation was selected. If errors exceeded 1°, Labeler C reviewed the task. If Labeler C’s annotations differed by less than 1° from either Labeler A or B, Labeler C’s annotations were accepted. Otherwise, Labeler D’s annotation was considered final. An example of images and their annotations from HVAngleEst is shown in Fig. 3.
The workflow for HVA, IMA, and line segments labeling: (1) Two parallel lines (red dashed lines) were drawn to annotate the great toe, the first metatarsal, and the second metatarsal. (2) The midpoints of these parallel lines were joined to create a straight line. (3) The points where this straight line intersects the edge of the phalanx defined the line segment points for that phalanx. The points (a, b), (c, d), and (e, f) correspond to the line segments of the great toe, the first metatarsal, and the second metatarsal. (4) HVA and IMA were calculated by the endpoints of each phalanx.
Examples of annotated images of one foot (top) and two feet (bottom). On the left is the de-identified original image; in the middle are the annotated boxes and line segments; and on the right is the fused image with the annotation results. The foot in the top image is labeled as “truncated” due to part of the foot being obstructed by the leg.
Data Records
The HVAngleEst dataset is publicly available for download via Science Data Bank (https://www.scidb.cn/en/s/FVFFnq)19 and can be accessed without registration. Figure 4 presents the dataset’s folder structure and file formats in detail.
The root folder of the dataset is named “HVAngleEst”. It contains subfolders named “images”, “annotations”, “tools”, as well as a “datasets.csv” file. Figure 4 provides an overview of the folder structure.
The CSV file “datasets.csv” contains the following information. “image_id” is the unique identifier of the image; “patient_id” refers to the patient to whom the image belongs; ‘filename’ is the name of the X-ray image; “source” indicates the image source, either from DICOM database or camera; “image_width” and “image_height” are the width and height of the image; “boxes” provides the coordinates of the box in format of “XMin, YMin, XMax, Ymax”, which are expressed in normalized image coordinates; “labels” specifies the object category, specifying either “left” or “right.”; “properties” indicates whether the foot is truncated; “great_toe”, “first_metatarsal”, and “second_metatarsal” denote coordinates of the two endpoints for each phalanx, formatted as “X1, Y1, X2, Y2”, are also expressed in normalized image coordinates; “HVA” and “IMA” are the hallux valgus angle and the intermetatarsal angle.
The “images” folder contains radiographs of feet. Each image filename begins with “IMG” followed by a zero-padded six-digit unique identifier and ends with the “.jpg” extension.
The “annotations” folder includes two subfolders: “boxes” and “masks”. The “boxes” folder contains annotation files for the localization task, organized into three subfolders: “COCO,” “YOLO,” and “PASCAL VOC.” The “COCO” folder includes a “json” file “COCO_feet_det.json” formatted according to the COCO20 standard. The “YOLO” and “PASCAL VOC” folders contain “.txt” and “.xml” files, respectively, which are named after the corresponding image files and follow the standard data formats of YOLO21 and PASCAL VOC22. Additionally, the “YOLO” folder includes a “classes.txt” file listing the available categories for localization, and there are two categories named “left” and “right”; The “masks” folder contains “.png” files, named after the corresponding image files. Based on the endpoints we annotated, these line segment masks were generated with a line width of 4 which was experimentally validated to yield the best hallux valgus angle estimation results15,16. The values assigned to the great toe, the first metatarsal, and the second metatarsal were 1, 2, and 3, respectively, while the background was assigned a value of 0.
The “tools” folder contains Python notebooks designed for support dataset preparation and algorithm implementation. During the dataset preparation phase, the notebook “points2angle.ipynb” was used to calculate HVA and IMA from the annotated endpoints of the phalanxes. The notebook “data2yolo.ipynb” generated ground truth annotations in YOLO format for localization, while “data2mask.ipynb” was used to create customized line segments based on the annotated phalanxes endpoints. Additionally, the YOLO format ground truth was converted into either the PASCAL VOC or COCO format using the notebooks “yolo2voc.ipynb” or “yolo2coco.ipynb”, respectively. During the linear regression stage, “seg2angle.ipynb” was employed to compute HVA and IMA from the output of the segmentation model.
Under the “splits” folder, there are 3 CSV files named “train.csv”, “val.csv” and ‘test.csv”. Each of these files contains a list of images used for training, validation and test in the technical validation.
Technical Validation
To ensure the dataset’s suitability for training deep learning algorithms, both foot localization and segmentation tasks were trained using YOLOv11s23 and DeeplabV324 based MobileNetV325 respectively, and a linear regression module to estimate HVA and IMA.
The dataset was randomly divided into 70% (967) for training, 20% (275) for validation, and 10% (140) for testing, for both the localization, segmentation models, as well as linear regression task. The training was performed on a Windows system equipped with an NVIDIA GeForce RTX 4060Ti GPU with 16 GB of VRAM. The localization model was pre-trained using YOLOv11s on the COCO dataset, while the segmentation model was pre-trained using MobileNetV3 on ImageNet26. The localization model was trained for 50 epochs, whereas the segmentation model was trained for 200 epochs. For the localization task, the input size was 640. For the segmentation task and Linear Regression, each foot was cropped using the bounding box, and the shorter side was resized to 512.
Localization performance
For the localization task, the feet were detected with a precision of 0.997, a recall of 1.0, and a mAP (mean Average Precision) of 0.994 at an IoU threshold of 0.5 on the test set.
Segmentation performance
For the segmentation task, line segments of the great toe, the first metatarsal, and the second metatarsal were predicted with mean mIoU (mean Intersection over Union) of 0.538 and pixel accuracy of 0.993 on the test set. Table 2 presents the mIoU for each category.
Linear regression performance
Table 3 presents the percentages of HVA and IMA with errors less than 3° and 5° on the test set. For errors less than 3°, the percentages were 94.4% for HVA and 79.4% for IMA. For errors less than 5°, the percentages were 96.9% for HVA and 93.8% for IMA.
Future research could compare the accuracy of weightbearing CT scans and X-rays in evaluating hallux valgus deformity, thereby providing more evidence-based guidance for clinical decision-making27,28,29.
Code availability
The Jupyter notebooks for dataset preparation and algorithm implementation are detailed in the Methods section. These Jupyter notebooks, along with the dataset, are available in the ‘tools’ folder at Science Data Bank (https://www.scidb.cn/en/s/FVFFnq)19. The code for technical validation can be accessed at (https://github.com/xahhyyWangQiong/HVAngleEst.git).
Change history
20 August 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41597-025-05820-0
References
Nishimura, A. et al. Prevalence of hallux valgus and risk factors among Japanese community dwellers. Journal of Orthopaedic Science. 19, 257–262, https://doi.org/10.1007/s00776-013-0513-z (2014).
Nix, S., Smith, M. & Vicenzino, B. Prevalence of hallux valgus in the general population: a systematic review and meta-analysis. Journal of foot and ankle research. 3, 1–9, https://doi.org/10.1186/1757-1146-3-21 (2010).
Fukushi, J.-i. et al. Comparison of outcomes of different osteotomy sites for hallux valgus: A systematic review and meta-analysis. Journal of Orthopaedic Surgery. 30(2), 10225536221110473, https://doi.org/10.1177/10225536221110473 (2022).
Mann, R. A. & Coughlin, M. J. Hallux valgus—etiology, anatomy, treatment and surgical considerations. Clinical Orthopaedics and Related Research®. 157, 31–41 (1981).
Vanore, J. V. et al. Diagnosis and treatment of first metatarsophalangeal joint disorders. Section 1: Hallux valgus. The Journal of Foot and Ankle Surgery. 42(3), 112–123, https://doi.org/10.1016/s1067-2516(03)70014-3 (2003).
Coughlin, M. J. & Freund, E. The reliability of angular measurements in hallux valgus deformities. Foot & ankle international. 22(5), 369–379, https://doi.org/10.1177/107110070102200503 (2001).
Coughlin, M. J., Saltzman, C. L. & Nunley, J. A. Angular measurements in the evaluation of hallux valgus deformities: a report of the ad hoc committee of the American Orthopaedic Foot & Ankle Society on angular measurements. Foot & ankle international. 23(1), 68–74, https://doi.org/10.1177/107110070202300114 (2002).
Azad, R. et al. Medical image segmentation review: The success of u-net. IEEE Transactions on Pattern Analysis and Machine Intelligence. https://doi.org/10.1109/TPAMI.2024.343557 (2024).
Chen, X. et al. Recent advances and clinical applications of deep learning in medical image analysis. Medical image analysis. 79, 102444, https://doi.org/10.1016/j.media.2022.102444 (2022).
Ju, R.-Y. & Cai, W. Fracture detection in pediatric wrist trauma X-ray images using YOLOv8 algorithm. Scientific Reports. 13(1), 20077, https://doi.org/10.1038/s41598-023-47460-7 (2023).
Ramesh, K., Kumar, G. K., Swapna, K., Datta, D. & Rajest, S. S. A review of medical image segmentation algorithms. EAI Endorsed Transactions on Pervasive Health & Technology. 7(27) https://doi.org/10.4108/eai.12-4-2021.169184 (2021).
Van den Borre, I. et al. Development and validation of a fully automated tool to quantify 3D foot and ankle alignment using weight-bearing CT. Gait & Posture. 113, 67–74, https://doi.org/10.1016/j.gaitpost.2024.05.029 (2024).
Xu, J., Jing, M., Wang, S., Yang, C. & Chen, X. A review of medical image detection for cancers in digestive system based on artificial intelligence. Expert review of medical devices. 16(10), 877–889, https://doi.org/10.1080/17434440.2019.1669447 (2019).
Kwolek, K., Liszka, H., Kwolek, B. & Gądek, A. Measuring the angle of hallux valgus using segmentation of bones on x-ray images. 313–325 Springer (2019).
Takeda, R. et al. Automatic estimation of hallux valgus angle using deep neural network with axis-based annotation. Skeletal Radiology. 53(11), 2357–2366, https://doi.org/10.1007/s00256-024-04618-2 (2024).
Xu, N., Song, K., Xiao, J. & Wu, Y. A Dataset and Method for Hallux Valgus Angle Estimation Based on Deep Learing. IEEE 34–40 (2022).
Tobin, J. et al. Domain randomization for transferring deep neural networks from simulation to the real world. IEEE; 23–30 (2017).
Russell, B. C., Torralba, A., Murphy, K. P. & Freeman, W. T. LabelMe: a database and web-based tool for image annotation. International journal of computer vision. 77, 157–173, https://doi.org/10.1007/s11263-007-0090-8 (2008).
Wang, Q. et al. HVAngleEst: A Dataset for End-to-end Automated Hallux Valgus Angle Measurement from X-Ray Images. https://doi.org/10.57760/sciencedb.19819 (2025).
Lin, T.-Y. et al. Microsoft coco: Common objects in context. 740–755 Springer (2014).
Redmon, J., Divvala, S., Girshick, R. & Farhadi, A. You only look once: Unified, real-time object detection. 779–788 (2016).
Everingham, M., Van Gool, L., Williams, C. K., Winn, J. & Zisserman, A. The pascal visual object classes (voc) challenge. International journal of computer vision. 88, 303–338 (2010).
Khanam, R. & Hussain, M. Yolov11: An overview of the key architectural enhancements. arXiv preprint arXiv:241017725 (2024).
Chen, L. et al. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv: Computer Vision and Pattern Recognition, 2017.
Howard, A.et al. Searching for mobilenetv3. IEEE; 1314–1324 (2019).
Deng, J. et al. Imagenet: A large-scale hierarchical image database. IEEE; 248–255 (2009).
Day, J. et al. A case-control study of 3D vs 2D weightbearing CT measurements of the M1-M2 intermetatarsal angle in hallux valgus. Foot & ankle international. 43(8), 1049–1052, https://doi.org/10.1177/10711007221091812 (2022).
Kvarda, P. et al. High reliability for semiautomated 3D measurements based on weightbearing CT scans. Foot & ankle international. 43(1), 91–95, https://doi.org/10.1177/10711007211034522 (2022).
Li, J. et al. Diagnostic applications and benefits of weightbearing CT in the foot and ankle: a systematic review of clinical studies. Foot and Ankle Surgery. 30(1), 7–20, https://doi.org/10.1016/j.fas.2023.09.001 (2024).
Acknowledgements
We extend our gratitude to all colleagues, visiting doctors, and students who contributed to processing portions of the HVAngleEst dataset over the past years.
Author information
Authors and Affiliations
Contributions
Qiong Wang: Conceptualization, processed the data, annotated the data, trained models, and wrote the paper. Dongdong Ji: Collected the data, cropped the data, and annotated the data. Junhu Wang: Annotated the data. Xinquan Yang, Peilong Liu, Liang Liu: collected the data. Yan Zhang, Jinqi Liang: Reviewed the experimental results. Hongmou Zhao: The project leader, annotated the data, revised the paper, and evaluated the experimental results.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Q., Ji, D., Wang, J. et al. HVAngleEst: A Dataset for End-to-end Automated Hallux Valgus Angle Measurement from X-Ray Images. Sci Data 12, 915 (2025). https://doi.org/10.1038/s41597-025-05261-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05261-9