Abstract
The Indo Pacific Convergence Zone (IPC) is recognized as an area of highest taxonomic and functional diversity. The polychaetes have garnered attention as their enormous functional trait diversity and their significant role in biomonitoring ecosystems. Here, we developed a comprehensive dataset for polychaetes in the IPC, encompassing information on species distribution, DNA barcodes, and functional traits. The species distribution data were collected from 39,310 occurrence records, with over 13% newly contributed from museum collections and 350 scientific papers. This dataset also provides 29% known species coverage in the IPC, 154 mitochondrial genomes (35.1% are new contributions), and 7,052 COI/18S/16S sequences (4.6% are new contributions). Our functional traits subdatabase comprises approximately 12,000 records and 2,831 species, belonging to 696 genera and 75 families, with 90% of which are new additions. The functional trait data were categorized into 13 traits spanning morphology, life history, physiology, and behavior. The datasetbase provide an unprecedented and original contribution for conservation and for studying the biogeography, evolution processes, and ecology of tropic organisms.
Similar content being viewed by others
Background & Summary
The Indo-Pacific Convergence Zone (IPC) is widely acknowledged as the highest marine biodiversity region, distinguished by its abundant biological resources. This extraordinary biodiversity has long captured the attention of biologists, with many species having been reported for their biodiversity, including coral, reef fishes, algae, snails, shellfish, lobsters and sea urchins1,2,3,4,5. Despite the immense importance of IPC in marine biodiversity, only a few datasets covering phylogenetic, functional diversity, and DNA barcode exist. On the other hand, while public databases such as the Global Biodiversity Information Facility (GBIF), Ocean Biodiversity Information System (OBIS), GenBank, and the Barcode of Life Data System (BOLD) deliver extensive data on species occurrences and DNA barcodes, significant challenges remain in the systematic consolidation and verification of this information. Consequently, the development of a comprehensive biodiversity database is essential to address these challenges effectively. Currently, only a limited number of well-studied animal groups, such as birds, mammals, and amphibians, have comprehensive databases that have been developed by researchers or institutions. These databases provide essential tools for research in biogeography, functional diversity, macroecology, macroevolution, and conservation. Unfortunately, comprehensive datasets on marine biodiversity, particularly within the IPC region, are rare.
The diversity of polychaetae and their functional traits make them an ideal model group for study the biodiversity and evolutionary processes of the IPC biodiversity center, as they have an intermediate lifespan and a diverse array of functional traits that highly adapted the local environment. Polychaeta is the most diverse and abundant class within the Phylum Annelida, with over 12,000 species documented6. These organisms are vital components of benthic fauna and ecosystems, playing crucial roles in bioturbation, nutrient cycling, and organic matter decomposition7,8. Furthermore, polychaetes exhibit a range of complex and diverse traits, including segmentation, regeneration and various body plan9,10. These characteristics make them an excellent model group for explaining the different responses of species and communities to environmental changes. changes. Until now, there have been few comprehensive and systematic scientific studies11 on the biodiversity of IPC’s polychaete which results in underestimation of biodiversity.
Constructing a comprehensive and reliable dataset is essential for understanding the biodiversity of polychaetes in the IPC region. In this study, we have developed a database that encompasses 2,831 species of polychaetes from 39,310 records from the IPC. Additionally, we present the results of a large-scale effort to DNA barcode polychaetes in this region, which includes 3,973 COI sequences, 1,574 16S sequences, 1,505 18S sequences, and 154 mitochondrial genomes. We also collected 13 functional traits, classified into four primary categories—morphology, physiology, life history, and behavior—yielding a dataset that represents at least 12,000 records. Our database serves as an invaluable resource for enhancing our understanding of the species diversity and ecological functions of polychaetes in the IPC. This comprehensive polychaete database is poised to benefit a broad community engaged in conservation, biogeography, evolutionary processes, and ecology.
Methods
Geographic coverage
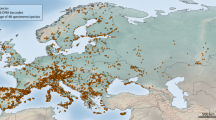
This study encompasses a geographic area that includes the Indian and Pacific Oceans, with longitudes ranging from 90°E to 180°E and latitudes extending from 30°N to 30°S, notably incorporating the renowned Coral Triangle (Fig. 1).
The workflow of the study is schematically represented in Fig. 2, illustrating each step from initial data collection to final integration and validation.
Occurrence of data collection and processing
Occurrence data for Polychaeta were collected from a variety of sources, including online databases (GBIF, OBIS), scientific literature, and field sampling. we conducted multiple intertidal zone collections and participated in deep-sea expedition samplings. All records obtained from diverse sources were consolidated into a single spreadsheet. Entries lacking species names, geographic coordinates, or fossil specimen records were removed. Additionally, we have updated species names utilizing the ‘Match Taxa’ tool from the World List of Marine Species (http://www.marinespecies.org)12. Due to the frequent conflict between the morphological definitions of hairy subspecies and molecular evidence, as well as the lack of standardized genetic thresholds, this database adopts a species-level classification to avoid taxonomic disputes. Only species identified by binomial nomenclature were retained, deliberately excluding subspecies classification. Geographic coordinates were converted to decimal format, and the ‘Round’ function was applied to refine the coordinates to four decimal places. Duplicates records with the identical species name, coordinates, depth, and collection time were eliminated. Furthermore, sampling sites were categorized by country/region based on georeferenced information, and the data is saved.
Functional traits collection and processing
Data were compiled data from a variety of publicly available sources, including the Polychaete Traits Database (http://polytraits.lifewatchgreece.eu/), the Chinese Animal Science Database (http://www.zoology.csdb.cn/), and Ocean Biodiversity Information System (https://obis.org/). Furthermore, we extracted and aggregated the data from hundreds of publications to further enrich our dataset. The traits identified were categorized into four primary categories: morphological traits, life history traits, physiological traits and behavioral traits, respectively. The specific criteria for categorization and coefficient settings are detailed in Table S1. When trait information exhibited variability among sources and was identified as representative of trait diversity, all pertinent datasets were included in the trait database.
DNA barcode data collection and processing
Three molecular markers-18S ribosomal RNA (18S rRNA), cytochrome c oxidase subunit I gene (COI), and 16S ribosomal RNA (16S rRNA)-as well as the mitochondrial genome, were employed to establish a DNA barcode database for Polychaeta in IPC. DNA barcode data were sourced from two primary avenues: sequence data were extracted from publicly accessible databases, such as GenBank and BOLD, while additional sequence information was generated through PCR experiments and whole genome sequencing (WGS). The specific methodologies employed in these processes are provided below:
-
(1)
Whole genomic DNA was extracted with the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocols. The fragments of COI, 16S, and 18S genes were amplified using Polychaeta COI, 16S, 18S primers (Table S2) and polymerase chain reaction (PCR), respectively. Amplifications were conducted in a reaction mixture containing 2 μl of template DNA, 12.5 μl of Premix TaqTM (Takara, Otsu, Shiga, Japan), 0.5 μl of each primer (10 mM), and double distilled H2O to a total volume of 25 μl. The cycling conditions were as follows: initial denaturation at 94 °C for 10 min, followed by 35–45 cycles of denaturation at 94 °C for 30 sec, annealing at 45 °C for 40 sec, and extension at 72 °C for 90 sec with a final extension at 72 °C for 5 min. PCR products were Sanger sequenced bidirectionally in Qingke Laboratory (Qingdao, China).
-
(2)
Sample tissues were subjected to whole genome sequencing (WGS) with a guarantee tha all samples originated from within the study area. The mitochondrial genomes of the species were subsequently assembled using software platforms such as GetOrganelle, MitoZ13,14. Finally, all mitochondrial genomes were independently reannotated using Mitos2 web tool15.
The relevant command parameters are as follows:
-
(1)
get_organelle_from_reads.py -1 sample_1.fastq -2 sample_2.fastq -R 80 -k 21,45,65,85,105,107,121,127 -t 16 -w 0.65 -F animal_mt -o animal_mt_out
-
(2)
mitoz all--thread_number 16--clade Annelida-segmented-worms--genetic_code 5--fq1 sample_1.fastq--fq2 sample_2.fastq--fastq_read_length 150--data_size_for_mt_assembly 3,0--assembler megahit--kmers_megahit 21 29 39 59 79 99 119 141--memory 50--requiring_taxa Annelida.
The initial sequence dataset contained a significant number of duplicates and sequences of suboptimal quality. To ensure accuracy, rigorous quality control measures were implemented during the sequence cleanup phase. Sequences containing non-ATGC bases were promptly eliminated. To eliminate redundancy across different sources, duplicate entries were identified and removed. We employed Seqkit2 to effectively utilize the rmdup function for sequence processing and used CD-HIT for clustering similar sequences16,17,18. Furthermore, we established stringent length screening criteria: 16S sequences were required to exceed 300 bp, 18S sequences had to be longer than 500 bp, and COI sequences needed to surpass 600 bp in length.
To detect and mark potential abnormal sequences in the system, this study employs a phylogenetic analysis approach. First, a global multiple sequence alignment is performed using MAFFT software19, with the G-INS-i algorithm selected to ensure high-accuracy sequence alignment. Subsequently, trimAI20 is applied for intelligent trimming of the alignment results, using the automated1 parameter to automatically optimize the sequences and eliminate the impact of low-confidence regions on subsequent analyses. Based on the trimmed dataset, a maximum likelihood phylogenetic tree is constructed using FastTree 221, with the GTR + T evolutionary model set to accurately capture the evolutionary relationships among the sequences. All abnormal sequences are highlighted in red in the“warning sequences” column.
Data Records
The dataset can be found in a figshare repository (Weng et al.22, and are licensed under CC BY.
The database, which includes three main components (occurrence records, DNA barcode data and functional traits), is organized into three distinct files formatted as ‘. Xlsx’. Instances of missing data within these files have been systematically designated as NA.
The “Occurrence_data.xlsx” file consists two sheets: one entitled ‘species distribution records’ and the other ‘species list’. Each record within the ‘species distribution records’ sheet contains detailed information, including the taxonomic category of the species, viz., family, genus, and species, as well as specific specimen details including latitude and longitude, date of collection, habitat type, depth, source of data, and country/region. Taxonomic category columns clarify the classification hierarchy, incorporating scientific names along with the author’s surname and the year of naming. Data sourced from public databases are marked with the respective database name, such as GBIF or OBIS, whereas literature-derived entries include the title of the publication. The depth column indicates the vertical water depth (in meters) where the species was found, and the habitat column characterizes the environments from which the specimens were collected. The ‘species list’ sheet provides taxonomic classifications for each species, including family, genus, and species.
The dataset comprises approximately 39,310 records of polychaete annelid worms, representing 2,831 species in 696 genera and 75 families, covering the period from 1776 to 2024. Notably, an 13% of these entries are derived from scientific literature, and it is important to highlight that this portion of the data is exclusive and not incorporated within existing databases. The majority of species records are marine, with a small number found in terrestrial or freshwater environments (Fig. 3). The period from 1991 to 2010 experienced the highest number of sampling events, totaling 12,089, which notably surpassed the 6,258 events documented in the decades from 1971 to 1990. Australia was identified as the country with the most sampling events, contributing 61.4% to the total, followed by Indonesia, China, India, and the Philippines. Australia was identified as the country with the most sampling events, contributing 61.4% to the total, followed by Indonesia, China, India, and the Philippines. Most sampling activities had occurred within the 0–100 meter. Furthermore, five families exhibiting the greatest species diversity include Syllidae, with 329 species; Nereididae, with 220 species; Terebellidae, with 215 species; Spionidae, with 174 species; and Polynoidae, with 142 species, as presented in Fig. 4.
The dataset titled “Functional_traits_data.xlsx” consists of a matrix comprising 2,831 species and 13 trait variables. A total of 11,953 valid trait recordings were collected, with temperature tolerance, salinity tolerance, depth zonation, and branching structure/branchiae being the four traits most frequently noted. Conversely, the traits with the fewest number of recordings were population spawning frequency, epistasis, and longevity (Fig. 5).
The dataset entitled “DNA_barcode_data.xlsx” consists of five separate sheets.
The first sheet, identified as “COI”, includes data relevant to the COI gene sequence, detailing information such as class, family, genus, and species, as well as the gene name (abbreviated as COI), gene length, GenBank ID, BOLD ID, and the nucleotide sequence.
The second sheet, labeled “16S”, contains information related to the 16S gene sequence, while the third sheet, named “18S”, provides analogous data for the 18S gene sequences. The columns in this sheet are consistent with those found in the COI sheet, thereby ensuring uniformity across the dataset. The fourth sheet, titled “mtDNA”, consists of mitochondrial genome data, featuring columns such as class, family, genus, species name, length, molecule type, GenBank ID, and sequence. Finally, the fifth sheet summarizes the gene collections affiliated with the species, containing four columns: COI, 16S, 18S, and mtDNA, where the values in the cells denote the number of sequences corresponding to each gene.
In the present study, we catalogued a total of 3,973 COI sequences, which accounts for 20.10% of the total species. Furthermore, we recorded 1,574 sequences for the 16S gene, corresponding to 17.20% of the species diversity. Moreover, we recorded 1,505 18S sequences, accounting for 20.28% of the overall species. In total, we also catalogued 154 mitochondrial genome sequences, of which 55 were generated in the present study. These sequences encompass 33 families, with Nereididae and Spionidae emerging as the most abundant (Fig. 6).
Statistics of DNA Barcode Data. (A) COI Gene: Depicts the proportion of species and the cumulative sequence count that incorporate the Cytochrome Oxidase I (COI) gene within the dataset. (B) 16S rRNA Gene: Displays the proportion of species and the total sequence number that harbor the 16S ribosomal RNA (16S rRNA) gene in the dataset. (C) 18S rRNA Gene: Illustrates the proportion of species and the aggregated sequence count that encompass the 18S ribosomal RNA (18S rRNA) gene in the dataset. (D) Mitochondrial Genome Distribution: Provides a statistical overview of the families represented, based on the presence of mitochondrial genomes in the dataset.
In the pie chart, the grey sections are species with sequences and the white sections are species without sequences.
Technical Validation
Occurrence data
We ensured the reliability of all occurrence records by verifying their origins through reputable databases and authoritative literature. Each extracted piece of information underwent a rigorous double-checking process, and all sources were meticulously documented. Species names were matched against the valid species lists from Worms, leading to the exclusion of any invalid or unknown species names from the database. Furthermore, geographical coordinates were scrutinized to confirm their alignment with the known distribution ranges of each species. Any outliers were rigorously investigated, corrected, or removed as necessary.
Functional trait data
For the collection of trait data, we utilized authoritative databases and publicly accessible journals. We recorded only those traits that were explicitly documented in the sources, avoiding any personal subjective interpretations. When conflicts arose between different sources, we performed an in-depth review of the textual information to resolve these discrepancies and maintain the accuracy of our dataset. We implemented a multi-step harmonization approach: (1) prioritize data with literature-verified specimens and standardized measurement protocols to minimize errors; (2) for highly conflicting traits with differences exceeding 20%, establish an expert panel to query additional information to resolve cases.
DNAbarcode data
Regarding gene sequence quality, we performed a rigorous quality check for each species. We employed computational tools to identify and eliminate sequences containing non-ATGC bases. Subsequently, we applied bioinformatics tools to eliminate redundancy, standardize the gene sequences, and enhance the overall quality and consistency. Given the varying publication dates of the data in public databases, we re-annotated all mitochondrial genomes using the MITOS2 software to ensure uniformity. Sequences with documented voucher specimens (e.g., museum IDs, ecological collection data) were prioritized to minimize misidentification risks from cryptic species or phenotypic convergence. Length/Ambiguity Filters: Sequences with abnormal lengths (e.g., COI <600 bp) or ambiguity (undetermined bases, “N”) were excluded to ensure alignment reliability. Phylogenetic Conflict Detection: Maximum Likelihood (ML) trees were constructed to identify sequences clustering anomalously (e.g., with distantly related taxa). Such sequences were flagged to alert users.
Usage Notes
Finally, we recommend that new users read and fully understand the relevant data files before use. We have provided the first extensive database of Polychaeta in IPC. If a user has an error to report or a suggestion to improve the database, the corresponding author can be contacted. Ultimately, this work aims to facilitate open-source data for both scientific and applied purposes. We currently maintain an irregular update schedule for the dataset, integrating new sampling and sequence data as they become available. To enhance efficiency and ensure timely updates, we are actively developing automated tools for data ingestion, validation, and integration.
Code availability
No custom code was used to generate or process the data described in this study.
References
Bellwood, D. R. & Meyer, C. P. Searching for heat in a marine biodiversity hotspot. Journal of Biogeography 36, 569–576 (2009).
Cowman, P. F., Parravicini, V., Kulbicki, M. & Floeter, S. R. The biogeography of tropical reef fishes: endemism and provinciality through time. Biological Reviews 92, 2112–2130 (2017).
Jorde, P. E., Andersson, A., Ryman, N. & Laikre, L. Are we underestimating the occurrence of sympatric populations? Molecular Ecology 27, 4011–4025 (2018).
Moore, A. M. et al. Unexpected discovery of diadema clarki in the coral triangle. Marine Biodiversity 49, 2381–2399 (2019).
Roberts, C. M. et al. Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 295, 1280–4 (2002).
Pamungkas, J., Glasby, C. J. & Costello, M. J. Biogeography of polychaete worms (annelida) of the world. Marine Ecology Progress Series 657, 147–159 (2021).
Sanchis, C., Soto, E. H. & Quiroga, E. The importance of a functional approach on benthic communities for aquaculture environmental assessment: trophic groups – a polychaete view. Marine Pollution Bulletin 167, 112309 (2021).
Welsh, D. T. It’s a dirty job but someone has to do it: the role of marine benthic macrofauna in organic matter turnover and nutrient recycling to the water column. Chemistry and Ecology 19, 321–342 (2003).
Kostyuchenko, R. P. & Kozin, V. V. Comparative aspects of annelid regeneration: towards understanding the mechanisms of regeneration. Genes 12, 1148 (2021).
Rieger, R. M. & Purschke, G. The coelom and the origin of the annelid body plan. Hydrobiologia 535, 127–137 (2005).
Faulwetter, S. et al. Polytraits: a database on biological traits of marine polychaetes. Biodiversity Data Journal 2, e1024 (2014).
Ahyong, S. et al. World register of marine species (WoRMS), WoRMS Editorial Board. (2024).
Jin, J. et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology 21, 241 (2020).
Meng, G., Li, Y., Yang, C. & Liu, S. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Research 47, e63 (2019).
Donath, A. et al. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res 47, 10543–10552 (2019).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Shen, W., Le, S., Li, Y. & Hu, F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/q file manipulation. PLoS One 11, e0163962 (2016).
Shen, W., Sipos, B., Zhao, L. SeqKit2: a swiss army knife for sequence and alignment processing. iMeta 3 (2024).
Rozewicki, J., Li, S., Amada, K. M., Standley, D. M. & Katoh, K. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Research 47, W5–W10 (2019).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Price, M. N., Dehal, P. S., Arkin, A. P. & Lawrence Berkeley National Laboratory Lbnl, B. C. U. S. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS one 5, e9490–e9490 (2010).
Weng, J. et al. Comprehensive Dataset for Polychaetes in the IPC: Species Distribution, DNA Barcodes, and Functional Traits, figshare, https://doi.org/10.6084/m9.figshare.27828573 (2025).
Acknowledgements
This research was supported by National Natural Science Foundation of China (42090044), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB42000000), National Natural Science Foundation of China (42376092), and the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (2022QNLM030004).We also thank Oceanographic Data Center and the high performance computer center, IOCAS, and China Science and Technology Cloud for support of data analysis. We thank the vessels of “KEXUE” for help in sample collection.
Author information
Authors and Affiliations
Contributions
L.L.Z. conceived the idea of the database and designed the study. J.Y.W., F.K.D., Q.Li, T.T.W.and C.R.L. gathered the data and performed data validation. W.X.W., C.R.L.,Y.W. and L.J. collected the samples and provided information about the habitats of the specimens in field. W.X.W. performed the morphological characterization and described the species. J.Y.W. wrote the first draft version of the manuscript. Y.N.L. was responsible for editing of the drafts of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Weng, J., Wu, X., Li, C. et al. Comprehensive Dataset for Polychaetes in the IPC: Species Distribution, DNA Barcodes, and Functional Traits. Sci Data 12, 956 (2025). https://doi.org/10.1038/s41597-025-05298-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05298-w