Abstract
Vestimentiferan tubeworms are representative species in deep-sea chemosynthetic ecosystems. Unlike most animals, these invertebrates lack mouths and digestive systems. Instead, their plumes function as the primary organ for direct metabolic exchange with the environment. Here, we present the single-cell transcriptome atlas of the Paraescarpia echinospica plume, offering single-cell resolution analysis for a deep-sea tubeworm. Our analysis revealed six cell clusters, namely hemocytes, proliferative cells, muscle cells, epithelial cells, nerve1 cells, and nerve2 cells, and we further characterized genes associated with immunity and transport. These findings establish a crucial foundation for future single-cell studies of tubeworms.
Similar content being viewed by others
Background & Summary
Cold seeps are regions where methane and hydrogen sulfide-rich fluids ascend from the seafloor at rates varying from a few centimeters to several meters annually1,2,3. Despite the challenging environmental conditions of darkness, extreme pressure, and toxicity, these ecosystems act as significant biodiversity hotspots, offering unique ecological niches that differ markedly from the surrounding seafloor4,5. These ecosystems support a large amount of invertebrates that form symbiotic relationships with chemoautotrophic bacteria, enabling survival in toxic environments6,7. Research into the invertebrates of cold seeps has significantly enhanced our comprehension of the deep-sea environment and the origins of life under extreme conditions.
Among these invertebrates, vestimentiferan tubeworms dominate cold-seep communities and serve as model organisms for studying adaptations to chemosynthetic ecosystems8,9. Vestimentiferan tubeworms lack digestive systems but possess a trophosome hosting sulfur-oxidizing bacteria, which sustain them through chemosynthesis10. Simultaneously, tubeworms supply symbiotic bacteria with essential nutrients such as carbon dioxide, oxygen, hydrogen sulfide via the plume to the trophosome11,12. The branchial plume, a highly vascularized organ analogous to gills, directly interfaces with seawater, mediating gas exchange and nutrient transport to the symbionts. The hemoglobin-rich blood vessels of plume exhibit an extraordinary affinity for toxic sulfides, a key adaptation to cold-seep environments13,14,15,16.
However, this exposure subjects the plume to constant microbial invasion17, necessitating robust immune defenses. While bulk transcriptomic studies of tubeworms have revealed elevated immune gene expression in the plume, these approaches obscure cell-type-specific responses18. Notably, Toll-like receptors (TLRs) and pathogen-defense genes are upregulated in the plume, but their spatial distribution have not been mapped to discrete cell types. Single-cell RNA sequencing (scRNA-seq) is essential to resolve this complexity, enabling identification of specialized cell subpopulations and their molecular mechanisms.
Paraescarpia echinospica, a species of vestimentiferan, is commonly found in numerous cold seeps situated in the western Pacific Ocean19,20,21. Previous studies have successfully decoded the genome sequence of P. echinospica22,23. In this study, scRNA-seq was employed to comprehensively characterize the cell types in the plume of P. echinospica. We captured a total of 10,689 high-quality cells and identified six main cell types based on their gene expression. Additionally, given the crucial roles of the plume in transport and immune response, we further characterized the status of transporters and immune genes. This work will establish a foundational resource for deep-sea organism studies at cellular resolution.
Methods
Sample collection
In brief, individuals of the deep-sea tubeworm P. echinospica were collected from the Haima cold seep (16°72.90′ N, 110°47.26′ E, 1387.1 m) in the South China Sea via the human occupied vehicle Shenhaiyongshi in February 2023. During the cruise, specimens were collected using a handnet and subsequently housed in a biobox. Immediately upon arrival at the main deck of the research vessel, one specimen was dissected, followed by rapid transfer of the P. echinospica plume to petri dishes for single-cell preparation in the onboard laboratory. The remaining tissues were immediately frozen and stored in a −80 °C freezer.
RNA extraction, library construction, and sequencing
Total RNA was extracted from the plume using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The concentration of RNA was determined using the NanoDrop (Thermo Fisher Scientific), and its integrity was assessed through the RNA Nano 6000 Assay Kit and Agilent 2100 Bioanalyzer (Agilent Technologies). Subsequently, an experimental process was conducted in accordance with the standard protocol provided by Oxford Nanopore Technologies (ONT). This process involved detecting the quality of the samples, constructing libraries, and assessing the quality of the libraries. Finally, the Nanopore PromethION platform was utilized to sequence the final library.
Transcriptome annotation
Initially, the original fastq data underwent filtration based on short fragments and low-quality reads (less than 500 bp in length and a Qscore < 6) to obtain a cleaned dataset. The complete sequences were then aligned to the previous published reference genome23 using the minimap2 software. Subsequently, clustering was performed based on comparison information, and consensus sequences were generated using the pinfish software. To ensure there were no duplicated transcript copies, a single consensus sequence was selected for each transcript. These consensus sequences were then merged and aligned with the reference genome using minimap2, and any redundant comparison results were removed. Afterward, transcripts with an identity below 0.9 and coverage less than 0.85 were filtered out, resulting in a final collection of 22,994 non-redundant transcript sequences.
Fresh tissues dissociated into single cell
Prior to library construction, the dissected plume of P. echinospica was transferred onto petri dishes and rinsed with ice-cold artificial seawater, prepared according to previous reports24. Then the plume was transferred on new petri dishes and minced by scalpel. The minced plume was collected into cell dissociation buffer containing 10 mg/ml Bacillus licheniformis protease (Sigma-Aldrich), the reaction was lasted for 30 minutes at 8 °C. After the reaction, the suspension was filtered through 30 μm cell strainer and spanned down at 350 g for 5 min at 4 °C. The supernatant was carefully removed, then the pellets were washed by ice-cold artificial seawater. After that, the pellets were resuspended in 1 × PBS containing 0.04% bovine serum albumin. The cell concentration was counted with Trypan Blue for subsequent DNBelab C Series Single-Cell library construction.
Single cell RNA-seq with DNBelab C4 system
The DNBelab C Series Single-Cell Library Prep Set (MGI) was employed for the preparation of barcoded scRNA-seq libraries25. Indexed sequencing libraries were then constructed following the manufacturer’s instructions and sequenced using the high-throughput DNBSEQ sequencer at the China National GeneBank (CNGB). The sequencing reads were generated in a paired-end format, with read 1 containing 30 bases that encompassed a 10 bp cell barcode 1, 10 bp cell barcode 2, and a 10 bp unique molecular identifier (UMI), and read 2 containing 100 bases representing the transcript sequence, along with a 10 bp sample index.
scRNA-Seq Data Processing and cell clustering
The raw data’s cell barcodes and UMIs were merged into fastq files using the parse feature in PISA (v1.10.2), and FastQC (v0.11.3) was utilized for quality control. After quality control, the raw reads were aligned to the reference genome of P. echinospica. We resequenced the plume’s transcriptome to improve gene annotation and adjusted the Unique Molecular Identifier (UMI) for sequencing errors. Gene matrices were generated using valid barcodes identified by the EmptyDrops method. These matrices were then imported into Seurat (v4.3.0) to perform cell clustering based on a graph-based approach. For cell filtration, cells with less than 200 genes or more than 5,000 genes in the gene cell barcode matrix were filtered out. Additionally, cells with mitochondrial UMIs accounting for more than 20% were also excluded. After performing PCA (30 PCs) and UMAP (default parameters) analysis on the integrated and batch corrected data, we proceeded to use the ‘findallmarker’ function (with only. pos equal to TRUE, min. pct and logfc. threshold equal to 0.25) to find differentially expressed genes (DEGs) between different clusters.
Data Records
The raw sequencing data (scRNA-seq and ONT transcriptomic sequencing data) reported in this paper are available at the China National GenBank (CNGB) Sequence Archive (CNSA, https://db.cngb.org/cnsa/) under accession number CNP000560726. All raw sequencing data were also deposited to the National Center for Biotechnology Information Gene Expression Omnibus database under the accession number: GSE28193127, GSE28333128.
Technical Validation
On board, the tubeworm was dissected for its plume, and the single-cell libraries of this tissue were constructed. These libraries were subsequently sequenced using the MGI DNB-seq platform in the laboratory. The scRNA-Seq data was processed using PISA and FastQC for quality control, and then mapped to the reference genome of P. echinospica. Cell clustering was performed using Seurat based on a graph-based approach, with cell filtration based on gene counts and mitochondrial UMIs. Furthermore, differential gene expression (DGE) analysis was conducted utilizing the ‘findallmarker’ function in Seurat (Fig. 1).
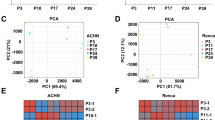
A total of 9,075,171,350 reads were generated from the raw data across seven libraries (Table 1). After quality control and raw data filtering, the genome mapping rates for these libraries are over 90%, indicating adequate sequencing quality for further data analysis. On average, each cell contained approximately 500 median genes (Fig. 2a), ~2,800 median UMI counts per cell (Fig. 2b), with ~13,000 total genes detected at an average sequencing depth of ~22,000 reads per cell. The sequencing saturation percentages ranged between 37.14% and 71.52% (Table 2). In most cells, the percentage of mitochondrial genes was less than 20% (Fig. 2c). Quality control of the seven libraries demonstrated no significant batch-specific differences among them (Fig. 2d).
Overview of quality control and cell filtering in each single-cell RNA sequencing library. (a) The violin plot of gene numbers in seven libraries after data filtering. (b) The violin plot of gene counts in seven libraries after data filtering. (c) Distribution of mitochondrial gene percentage in each library after data filtering. (d) The distribution of seven libraries in UMAP.
After quality control, a total of 10,689 high-quality cells were obtained. We successfully annotated six cell types, including hemocytes, proliferative cells, muscle cells, epithelial cells, nerve1 cells, and nerve2 cells (Fig. 3a). The genes that distinguish these clusters were visualized in a heatmap, highlighting the top 50 differentially expressed genes (Fig. 3b and Supplemental Table S1). Furthermore, the expression levels of these marker genes were illustrated in DotPlot (Fig. 3c and Supplemental Table S2). Our single-cell data reveals that nerve cells (comprising both nerve1 cells and nerve2 cells) and muscle cells account for 29% and 24% of the total cell population, respectively, making them the main components of the plume (Fig. 3d). We subsequently investigated the correlation among different cell types, and the results show a strong correlation between muscle cells, epithelial cells and nerve cells (Fig. 3e).
Single-cell transcriptional landscape of tubeworm plume and cell cluster identification. (a) The analysis of 10,689 cells from the tubeworm using UMAP to visualize and color-code the cells according to their cluster cell type. Each cell is represented by a dot on the plot, and the different colors correspond to specific cell types. (b) Heatmap showing the expression of the top 50 DEGs of the main cell types. Colors range from yellow (low expression) to red (high expression). (c) DotPlot of marker genes in 6 different cell types. (d) The pie chart displays the proportion of each cell type out of the total number of cells. (e) The heatmap displays the similarity between cell types, with colors ranging from blue to red indicating high to low similarity, respectively.
We examined the expression patterns of transport proteins in different cell types and downloaded 1,801 transport-related genes from the “transporters” catalog in the KEGG BRITE Database (https://www.kegg.jp/brite/ko02000). Subsequently, these genes were screened among DEGs across all cell types, and 48 transport-related genes were identified (Fig. 4a and Supplemental Table S3). Notably, high expression of globin (glob1)29 and neuroglobin (ngb)30 involved in oxygen transport were found in hemocytes (Fig. 4b). High expression of slc6a8, a creatine transporter, was also discovered in hemocytes (Fig. 4b). Creatine, acting as a blood antioxidant, protects cells from oxidative damage and genotoxicity, and potentially extends the lifespan of hemocytes31. Presumably, the creatine transporter assists tubeworm hemocytes to better maintain their life activity and life cycle of tubeworm hemocytes in harsh environments. Furthermore, slc5a632, slc6a533 and dur334 are highly expressed in epithelial cells (Fig. 4c). Overall, the elevated expression of solute carrier (SLC) transporter in these cell types enables efficient molecule transportation across epithelial barriers and supports specialized cellular functions. Hemocytes are known as immune cells in invertebrates35. Notably, we investigated some immune-related genes from DEGs including mfge836, fut837 and plg38 in hemocytes (Fig. 4d). In summary, our study provides a valuable scRNA-seq dataset for investigating tubeworm gene expression profiles and offers an in-depth exploration of the molecular mechanisms underlying their adaptation to extreme environments.
scRNA analysis of transport and immune related genes. (a) Heatmap illustrates the expression levels of transport-related genes across different cell types, with each column representing a specific cell type. Colors range from blue (low expression) to red (high expression). (b) Violin plots of transport genes specially expressed in hemocytes. (c) Violin plots of transport genes specially expressed in epithelial cells. (d) Violin plots of immune genes in each cell type.
Usage Notes
All sequencing data has been deposited in the CNGB database and the GEO database, including the raw FASTQ files and the expression matrix files processed by DNBC4tools. We have also uploaded our scripts to GitHub (refer to the Code Availability section for more details), allowing other researchers to reproduce our results or integrate them with other datasets for further analysis.
Code availability
The scRNA-seq analysis scripts for this study have been uploaded to GitHub (https://github.com/littersix123/tubeworm_script).
References
Suess, E. Marine cold seeps and their manifestations: geological control, biogeochemical criteria and environmental conditions. International Journal of Earth Sciences 103, 1889–1916, https://doi.org/10.1007/s00531-014-1010-0 (2014).
Brown, K. M., Tryon, M. D., DeShon, H. R., Dorman, L. M. & Schwartz, S. Y. Correlated transient fluid pulsing and seismic tremor in the Costa Rica subduction zone. Earth and Planetary Science Letters 238, 189–203, https://doi.org/10.1016/j.epsl.2005.06.055 (2005).
Kennicutt, M. C. et al. Vent-type taxa in a hydrocarbon seep region on the Louisiana slope. Nature 317, 351–353 (1985).
Felden, J. et al. Anaerobic methanotrophic community of a 5346-m-deep vesicomyid clam colony in the Japan Trench. Geobiology 12, 183–199, https://doi.org/10.1111/gbi.12078 (2014).
Ruff, S. E. et al. Microbial communities of deep-sea methane seeps at Hikurangi continental margin (New Zealand). PLoS One 8, e72627, https://doi.org/10.1371/journal.pone.0072627 (2013).
Dubilier, N., Bergin, C. & Lott, C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nature Reviews Microbiology 6, 725–740 (2008).
Petersen, J. M. & Dubilier, N. Methanotrophic symbioses in marine invertebrates. Environmental Microbiology Reports 1, 319–335 (2009).
Sibuet, M. & Olu, K. Biogeography, biodiversity and fluid dependence of deep-sea cold-seep communities at active and passive margins. Deep Sea Research Part II: Topical Studies in Oceanography 45, 517–567 (1998).
Takishita, K., Kakizoe, N., Yoshida, T. & Maruyama, T. Molecular evidence that phylogenetically diverged ciliates are active in microbial mats of deep-sea cold-seep sediment. J Eukaryot Microbiol 57, 76–86, https://doi.org/10.1111/j.1550-7408.2009.00457.x (2010).
Nussbaumer, A. D., Fisher, C. R. & Bright, M. Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441, 345–348 (2006).
Minic, Z. & Hervé, G. Biochemical and enzymological aspects of the symbiosis between the deep‐sea tubeworm Riftia pachyptila and its bacterial endosymbiont. European Journal of Biochemistry 271, 3093–3102 (2004).
Girguis, P. R. et al. Fate of nitrate acquired by the tubeworm Riftia pachyptila. Applied and environmental microbiology 66, 2783–2790 (2000).
Arp, A. J. & Childress, J. J. Blood function in the hydrothermal vent vestimentiferan tube worm. Science 213, 342–344 (1981).
Zal, F., Lallier, F. H., Green, B. N., Vinogradov, S. N. & Toulmond, A. The Multi-hemoglobin System of the Hydrothermal Vent Tube Worm Riftia pachyptila: II. Complete Polypeptide Chain Composition Investigated by Maximum Entropy Analysis of Mass Spectra (∗). Journal of Biological Chemistry 271, 8875–8881 (1996).
Arp, A. J. & Childress, J. J. Sulfide binding by the blood of the hydrothermal vent tube worm Riftia pachyptila. Science 219, 295–297 (1983).
Terwilliger, R., Terwilliger, N., Bonaventura, C., Bonaventura, J. & Schabtach, E. Structural and functional properties of hemoglobin from the vestimentiferan Pogonophora, Lamellibrachia. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology 829, 27–33 (1985).
Ruff, S. E. et al. Global dispersion and local diversification of the methane seep microbiome. Proc Natl Acad Sci USA 112, 4015–4020, https://doi.org/10.1073/pnas.1421865112 (2015).
Yang, Y. et al. Genomic, transcriptomic, and proteomic insights into the symbiosis of deep-sea tubeworm holobionts. Isme j 14, 135–150, https://doi.org/10.1038/s41396-019-0520-y (2020).
Southward, E. C., Schulze, A. & Tunnicliffe, V. Vestimentiferans (Pogonophora) in the Pacific and Indian Oceans: a new genus from Lihir Island (Papua New Guinea) and the Java Trench, with the first report of Arcovestia ivanovi from the North Fiji Basin. Journal of natural History 36, 1179–1197 (2002).
Zhao, Y. et al. Ecological characterization of cold-seep epifauna in the South China Sea. Deep Sea Research Part I: Oceanographic Research Papers 163, 103361 (2020).
Feng, D. et al. Cold seep systems in the South China Sea: An overview. Journal of Asian Earth Sciences 168, 3–16 (2018).
Sun, Y. et al. The mitochondrial genome of the deep-sea tubeworm Paraescarpia echinospica (Siboglinidae, Annelida) and its phylogenetic implications. Mitochondrial DNA Part B 3, 131–132 (2018).
Sun, Y. et al. Genomic Signatures Supporting the Symbiosis and Formation of Chitinous Tube in the Deep-Sea Tubeworm Paraescarpia echinospica. Mol Biol Evol 38, 4116–4134, https://doi.org/10.1093/molbev/msab203 (2021).
Dickson, A. G. & Goyet, C. Handbook of methods for the analysis of the various parameters of the carbon dioxide system in sea water. Version 2. (Oak Ridge National Lab.(ORNL), Oak Ridge, TN (United States) 1994).
Liu, C. et al. A portable and cost-effective microfluidic system for massively parallel single-cell transcriptome profiling. BioRxiv, 818450 (2019).
Zhang, Y. et al. A single-cell plume transcriptome atlas of the cold-seep tubeworm Paraescarpia echinospica. CNGB. https://db.cngb.org/search/project/CNP0005607/ (2024).
Zhang, Y. et al. A single-cell plume transcriptome atlas of the cold-seep tubeworm Paraescarpia echinospica. GEO https://identifiers.org/geo/GSE281931 (2024).
Zhang, Y. et al. A single-cell plume transcriptome atlas of the cold-seep tubeworm Paraescarpia echinospica. GEO https://identifiers.org/geo/GSE283331 (2024).
Jourd’heuil, F. L., Xu, H. & Jourd’heuil, D. 337 - Cytoglobin Prevents Redox-Dependent Smooth Muscle Cell Apoptosis During Vascular Injury. Free Radical Biology and Medicine 100, S144, https://doi.org/10.1016/j.freeradbiomed.2016.10.378 (2016).
Mathai, C., Jourd’heuil, F. L., Lopez-Soler, R. I. & Jourd’heuil, D. Emerging perspectives on cytoglobin, beyond NO dioxygenase and peroxidase. Redox Biol 32, 101468, https://doi.org/10.1016/j.redox.2020.101468 (2020).
Mahmood, R. & Qasim, N. M. 24 - Protective effect of creatine against hyperglycemia induced oxidative damage in human erythrocytes. Free Radical Biology and Medicine 112, 32, https://doi.org/10.1016/j.freeradbiomed.2017.10.037 (2017).
de Carvalho, F. D. & Quick, M. Surprising substrate versatility in SLC5A6: Na+−coupled I- transport by the human Na+/multivitamin transporter (hSMVT). J Biol Chem 286, 131–137, https://doi.org/10.1074/jbc.M110.167197 (2011).
Marques, B. L. et al. Neurobiology of glycine transporters: From molecules to behavior. Neurosci Biobehav Rev 118, 97–110, https://doi.org/10.1016/j.neubiorev.2020.07.025 (2020).
Uemura, T., Kashiwagi, K. & Igarashi, K. Polyamine uptake by DUR3 and SAM3 in Saccharomyces cerevisiae. Journal of Biological Chemistry 282, 7733–7741 (2007).
Lavine, M. & Strand, M. Insect hemocytes and their role in immunity. Insect biochemistry and molecular biology 32, 1295–1309 (2002).
Fricker, M. et al. MFG-E8 mediates primary phagocytosis of viable neurons during neuroinflammation. J Neurosci 32, 2657–2666, https://doi.org/10.1523/jneurosci.4837-11.2012 (2012).
Nakayama, K. et al. Core fucose is essential glycosylation for CD14-dependent Toll-like receptor 4 and Toll-like receptor 2 signalling in macrophages. J Biochem 165, 227–237, https://doi.org/10.1093/jb/mvy098 (2019).
Plow, E. F., Ploplis, V. A., Busuttil, S., Carmeliet, P. & Collen, D. A role of plasminogen in atherosclerosis and restenosis models in mice. Thromb Haemost 82(Suppl 1), 4–7 (1999).
Acknowledgements
This work was financially supported by the National Key R&D Program of China (2022YFC2805404), the Major Scientific and Technological Projects of Hainan Province (ZDKJ2021028). The research was supported by the Project of Sanya Yazhou Bay Science and Technology City (SKJC-2024-01-002). This work was supported by High-performance Computing Platform of YZBSTCACC (YaZhou Bay Science and Technology City Advanced Computing Center). We also would like to express our sincerest gratitude to the pilots on HOV Shenhaiyongshi and crews on R/V Tansuoerhao for their professional service during the cruise of TS2-21 in 2023. We appreciate the technical support from China National GeneBank for the library sequencing.
Author information
Authors and Affiliations
Contributions
Design and conceptualization: L.M., S.L., H.Z.; Sample collection: N.Z., J.M., Y.X.; investigation: X.Z., Y.Z., N.Z., J.M., Q.J., Z.C., X.D.; Formal analysis and visualization: X.Z., H.C.; resources: S.L., H.Z.; Supervision: Z.Z., G.F., W.Z.; Writing original draft: Y.Z., X.Z., N.Z.; Writing review & editing: L.M., H.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, Y., Zhang, X., Zhang, N. et al. A single-cell plume transcriptome atlas of the cold-seep tubeworm Paraescarpia echinospica. Sci Data 12, 1020 (2025). https://doi.org/10.1038/s41597-025-05392-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05392-z
This article is cited by
-
A single-cell plume transcriptome atlas of the cold-seep tubeworm Paraescarpia echinospica
Scientific Data (2025)