Abstract
Clinical trials are pivotal for developing new medical treatments but typically carry risks such as patient mortality and enrollment failure that waste immense efforts spanning over a decade. Applying artificial intelligence (AI) to predict key events in clinical trials holds great potential for providing insights to guide trial designs. However, complex data collection and question definition requiring medical expertise have hindered the involvement of AI thus far. This paper tackles these challenges by presenting a comprehensive suite of 23 meticulously curated AI-ready datasets covering multi-modal input features and 8 crucial prediction challenges in clinical trial design, encompassing prediction of trial duration, patient dropout rate/event, serious adverse event, mortality event, trial approval outcome, trial failure reason, drug dose, and design of eligibility criteria. Furthermore, we provide basic validation methods for each task to ensure the datasets’ usability and reliability. We anticipate that the availability of such open-access datasets will catalyze the development of advanced AI approaches for clinical trial design, ultimately advancing clinical trial research and accelerating medical solution development.
Similar content being viewed by others
Background & Summary
The clinical trial process is an essential step in developing new treatments (e.g., drugs, vaccines, or medical devices), serving as the bridge between scientific discovery and real-world medical application. Clinical trials are designed to systematically evaluate the safety, efficacy (in treating specific diseases), dosage, and overall impact of these treatments on human bodies1. The basic steps of a clinical trial typically include: (1) planning and design, where researchers define the study objectives, eligibility criteria, and determine the treatment protocol; (2) recruitment and screening, where eligible participants are enrolled and baseline health data is collected; (3) intervention and monitoring, where participants receive treatment or placebo, and their health outcomes are closely monitored; (4) data analysis, where results are analyzed to determine the treatment’s safety and efficacy; (5) conclusion, findings are reported and, if successful, submitted for regulatory approval. Typically conducted in multiple phases (Phase 1 to Phase 4, approved by FDA after passing Phase 3), these trials begin with small-scale studies to assess safety and dosage (Phase 1, 20-80 healthy volunteers), expand to evaluate effectiveness and side effects in larger populations (Phase 2 and 3, 100-300 and 300-3,000 patients respectively), and continue into post-marketing surveillance to monitor long-term outcomes (Phase 4, several thousand to tens of thousands of patients)2. However, these exploratory trials have a high failure rate3,4. Compounding the issue, clinical trials are known for being time-consuming, labor-intensive, and costly. Clinical development programs containing the set of phase 1-3 trials typically span 7-11 years, cost an average of 2 billion USD, and achieve approval rates of only around 15%5. Clinical trials are inherently risky as they explore “new” treatments, while artificial intelligence (AI) is particularly well-suited for making accurate estimates to reduce risk since AI excels at identifying patterns, including those not previously known to humans6.
Years of clinical trials have generated a vast amount of multi-modal data7, encompassing aspects such as inclusion/exclusion criteria designs, adverse event statistics, and patient enrollment results. Such extensive data offers a robust foundation for developing advanced AI algorithms8. However, identifying key clinical trial challenges and effectively leveraging the complex variables within this data require a blend of deep medical knowledge and AI expertise. This complexity has hindered skilled AI experts from fully utilizing the data.
The ClinicalTrials.gov website (https://clinicaltrials.gov/) provides comprehensive information on clinical trials, including study protocols, participant eligibility criteria, and study results, making it a valuable resource for AI engineers and medical professionals. This centralized repository covers more than 480,000 clinical trial records (as of Feb 2024) from all 50 US states and international trials from 221 countries. However, identifying key clinical trial challenges suitable for AI solutions and selecting appropriate variables for different challenges remain problematic for data scientists who lack relevant background knowledge.
To facilitate cross-disciplinary research and fully leverage the expertise of data scientists and AI experts9,10, this paper identifies 8 key critical clinical trial challenges. It organizes 23 corresponding AI-ready datasets to support their involvement in these tasks. The data, representing clinical trials registered before February 16, 2024, were collected from ClinicalTrials.gov. We extracted elements and attributes from the XML records of each clinical trial and converted them into tabular data formats, which are better suited for processing by AI models, including deep learning models. Additionally, we transformed some features into more informative forms; for example, converting health condition information into ICD-10 codes. We also enrich our data with valuable information from DrugBank (e.g., drug molecular structures and pharmaceutical properties as feature)11 and TrialTrove (e.g., trial approval information as ground truth, https://pharmaintelligence.informa.com/products-and-services/data-and-analysis/trialtrove) to depict a comprehensive set of information for clinical trial AI. Our organized datasets are available at: https://huyjj.github.io/Trialbench/.
When curating these datasets, we manually determined the prediction objectives for each task and selected variables according to the timing of applying AI in real-world practice. For instance, we ensured that trial result information was not included if the AI task is to be performed before trial completion. Features with a limited number of discrete options were organized into categorical features. Each task ultimately has a clearly defined prediction objective and a collection of input tabular variables. Unlike traditional tabular datasets, these datasets may contain multi-modal input features, such as free text (e.g., eligibility criteria) and graph data (e.g., drug molecular graphs).
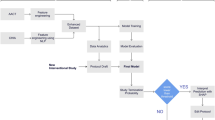
Figure 1 illustrates the TrialBench platform, containing 8 well-defined clinical trial design tasks. The TrialBench platform provides 23 corresponding AI-ready datasets across these 8 tasks, implemented evaluation metrics, and baseline models. AI experts can easily access the datasets and targets to develop advanced models, evaluate models on specific metrics, and compare them against baseline models for reference.
Overview of TrialBench. (a) TrialBench comprises 23 AI-ready clinical trial datasets for 8 well-defined tasks: clinical trial duration forecasting, patient dropout rate prediction, serious adverse event, all-cause mortality event prediction, trial approval outcome prediction, trial failure reason identification, eligibility criteria design, and drug dose finding. For each task, we extracted appropriate multi-modal variables and prediction targets from ClinicalTrials.gov, implemented evaluation metrics, and constructed a multi-modal baseline model to assess dataset quality and to serve as the baseline model. We integrate drug SMILES strings, textual descriptions (e.g., eligibility criteria), Medical Subject Heading (MeSH) term, disease ICD-10 code, and other categorical or numerical features as up to five distinct modal features. The multi-modal model utilizes message-passing neural networks (MPNNs), Bio-BERT, MeSH embedding layer, Graph-based Attention Model (GRAM), and DANet basic blocks to process each modality, respectively. (b) We present the trial failure reason identification task (a classification task) as an illustrative example to aid understanding, showcasing the input features, prediction target, baseline model, and some evaluation metrics. TP: True Positive; FP: False Positive; TN: True Negative; FN: False Negative; Prec: Precision.
Methods
AI-solvable Clinical Trial Task Definitions
In this paper, we identify 8 AI-solvable clinical trial tasks. For each task, we elaborate on its background, explain how it would help clinical trial design and management, curate the dataset, evaluate the performance of well-known AI methods, and report the empirical results. Table 1 summarizes and compares all the AI-solvable clinical trial tasks and corresponding datasets. We provide the following three aspects for each learning task: (1) Background. Background of the learning task. (2) Definition. A formal definition of the learning task (input feature and output). (3) Broad impact. The broader impact of advancing real clinical trials on the task.
Trial Duration Prediction
Background. The duration of a clinical trial is defined as the number of years from the trial’s start date to its completion date, representing a continuous numerical value. The clinical trial duration is directly related to its cost because longer trials require more extended use of resources, including personnel, facilities, and materials, leading to increased expenses12.
Definition. This task focuses on predicting trial duration (time span from the enrollment of the first participant to the conclusion of the study) based on multi-modal trial features such as eligibility criteria, target disease, etc. It is formulated as a regression task.
Broad impact. Predicting the duration of clinical trials offers several significant benefits that enhance drug development efficiency and effectiveness. AI-driven predictions allow for better planning and resource allocation, leading to more accurate staffing, budgeting, and management of clinical sites. This enhances decision-making by enabling stakeholders to prioritize projects based on expected timelines and identify risks early, allowing for proactive measures to mitigate delays. Ultimately, accurate duration predictions assists pharmaceutical companies in more accurately estimating costs, determining the right number of sites for potential acceleration, and strategizing effective market launch plans in a single, comprehensive solution.
Patient Dropout Prediction
Background. Prior studies have shown that approximately 30% of participants eventually drop out of clinical trials13, potentially undermining the validity of trial outcomes and contributing to higher costs and prolonged timelines.
Definition. This task seeks to predict both the occurrence (binary classification) and rate (regression) of patient dropout in clinical trials, based on multi-modal features such as eligibility criteria, target disease, and other protocol-level information. It is formulated as a dual-objective learning problem comprising a classification subtask for dropout occurrence and a regression subtask for dropout rate estimation.
Broad impact. Predicting patient dropout in clinical trials holds significant promise for improving the efficiency and effectiveness of drug development processes. Predicting patient dropout rates can improve the efficiency of clinical trials. High dropout rates often necessitate the recruitment of additional participants to meet the required sample size, which can be both time-consuming and costly.
Adverse Event Prediction
Background. Adverse event prediction is crucial in clinical trials as it directly impacts the safety, efficacy, and overall success of the trial. The primary concern in any clinical trial is the safety of the participants14.
Definition. The task targets predicting the occurrence of adverse events given multi-modal clinical trial features such as drug molecule, target disease, eligibility criteria, etc. It is formulated as a binary classification problem.
Broad impact. Predicting adverse events helps in identifying potential risks to patients before they occur, allowing for proactive measures to be taken. On the other hand, regulatory organizations such as the FDA and EMA have strict guidelines for monitoring and reporting adverse events in clinical trials15. Accurate prediction and early detection of adverse events can ensure compliance with these regulations.
Mortality Event Prediction
Background. The mortality event in a clinical trial refers to the death of participants during the study period. When serious adverse events escalate beyond a critical threshold, unsafe treatments or severe disease conditions may lead to fatalities. An unexpectedly mortality can trigger ethical concerns and necessitate a thorough safety reassessment16. As such, the occurrence of mortality events serves as a key indicator for evaluating the safety and potential risks associated with the treatment or intervention under investigation.
Definition. This task aims to predict the occurrence of mortality in a clinical trial based on multi-modal features, including drug molecules, target diseases, eligibility criteria, and others. It is formulated as a binary classification problem.
Broad impact. Accurate prediction of trial-related mortality enhances patient safety by enabling early identification of high-risk scenarios and timely intervention. It also informs more efficient trial designs, optimizing resource allocation and reducing overall costs. By accelerating drug development and improving regulatory compliance, such predictions contribute to faster delivery of effective treatments and reinforce public trust and ethical integrity in clinical research.
Trial Approval Prediction
Background. Clinical trial approval refers to whether a drug can pass a certain phase of clinical trial, which is the most important outcome of a clinical trial. Recent investigations suggest that clinical trial suffers from low approval rate17.
Definition. This task aims to predict the probability of trial approval given multi-modal trial features such as drug molecule, disease code, and eligibility criteria. It is formulated as a binary classification problem.
Broad impact. Predicting trial approval can enhance the efficiency and success rates of drug development. By accurately forecasting which drugs are likely to pass clinical trial phases, companies can focus their resources on the most promising candidates, reducing wasted time and money on less viable options. This targeted approach can accelerate the development of effective treatments, bringing them to market faster and improving patient outcomes. Additionally, reliable approval predictions can streamline regulatory processes and increase investor confidence in the pharmaceutical industry.
Trial Failure Reason Identification
Background. Clinical trials usually fail due to a couple of reasons18: (1) business decision (e.g., lack of funding, company strategy shift, pipeline reorganization, drug strategy shift); it is challenging to predict business decision, so we do not involve these trials in our dataset; (2) Poor enrollment. Insufficient enrollment can compromise the statistical power of the study, making it difficult to detect a significant effect of the drug. Also, poor enrollment can lead to delays in the trial timeline and increased costs, as more resources are required to recruit additional participants. (3) Safety. Unexpected adverse reactions or side effects can occur, posing significant risks to participants’ health. This can lead to the trial being halted or terminated. (4) Efficacy (effectiveness). In the trial, we expect the tested drug to outperform the standard treatment in curing the target disease. Thus, efficacy (effectiveness) is typically required.
Definition. Given clinical trial features, the goal of this task is to leverage the AI model to classify it into one of these four categories, including (1) successful trials, (2) failure due to poor enrollment, (3) failure due to drug safety issue; (4) fail due to lack of efficacy. It is a multi-category (4 categories) classification problem.
Broad impact. Accurately predicting the reasons for clinical trial failures can greatly enhance the efficiency of drug development by preventing costly delays and optimizing resource allocation. This leads to faster delivery of effective treatments to patients, improving patient outcomes and public health. Additionally, better-designed trials with higher success rates can encourage greater confidence and participation in clinical research.
Eligibility Criteria Design
Background. To achieve statistically significant results, a clinical trial must meet its target sample size19. Insufficient patient numbers can lead to underpowered studies, which may fail to demonstrate the effectiveness of a treatment or may miss important safety information. Eligibility criteria are essential to patient recruitment20. They describe the patient recruitment requirements in unstructured natural language. Eligibility criteria comprise multiple inclusion and exclusion criteria, which specify what is desired and undesired when recruiting patients. Each individual criterion is usually a natural language sentence.
Definition. This task aims to design eligibility criteria given a series of clinical trial features such as target disease, phase, drug molecules, etc.
Broad impact. Using AI models to design eligibility criteria for clinical trials offers several significant advantages. AI can predict which patients are more likely to meet the eligibility criteria based on historical data and real-world evidence. This speeds up the recruitment process by identifying suitable candidates faster and reducing the time and cost associated with screening large numbers of unsuitable participants.
Drug Dose Finding
Background. One of the primary goals of clinical trials is to determine the drug dose. Determining the correct dosage of a drug is crucial to ensure its effectiveness in treating a particular condition. In the early stages of drug development, predicting the optimal dosage is essential for designing clinical trials21,22.
Definition. This task aims to predict drug dosage based on drug molecular structure and target disease, which is formulated as an ordinal classification problem.
Broad impact. By estimating the dose-response relationship and identifying the dosage range that balances efficacy and safety, researchers can design more informative and efficient clinical studies.
Raw Data
Our primary data source is the clinicalTrials.gov website (https://clinicaltrials.gov/), which serves as a publicly accessible resource for clinical trial information. Supported by the U.S. National Library of Medicine, this database encompasses over 420,000 clinical trial records, spanning all 50 U.S. states and 221 countries worldwide. Table 2 The number of recorded trials would grow rapidly with time, as shown in Fig. 2(a). Table 3 reports some essential statistics of the curated datasets, including the number of involved trials, drugs, diseases, and proportion of interventional trials. There are hundreds of multi-modal features in ClinicalTrials.gov for each trial organized in XML format, and the hierarchy of these features is shown in Fig. S1. Table 2 demonstrates a real clinical trial example.
(a) A histogram showing the distribution of start dates for the selected trials reveals a steady increase in the number of initiated trials over time, reflecting the growing demand for new treatments. (b) A statistical breakdown of the clinical trials by phase indicates that the majority of trials are in Phase II. (c) The frequency of events varies across phases, as exemplified by the occurrence of participant dropout.
Data Acquisition
We create the dataset benchmark from multiple public data sources, including ClinicalTrials.gov, DrugBank, TrialTrove, ICD-10 coding system, as elaborated below.
-
ClinicalTrials.gov is a publicly accessible database maintained by the U.S. National Library of Medicine (NLM) at the National Institutes of Health (NIH). It provides detailed information about clinical trials conducted around the world, including those funded by public and private entities. Each clinical trial in ClinicalTrials.gov is provided as an XML file, which we parse to extract relevant variables. For each trial, we retrieve the NCT ID (unique identifiers for each clinical study), disease names, associated drugs, title, summary, trial phase, eligibility criteria, results of statistical analyses, other details, and then integrate into our data. Some of these features are not always available. For example, observational clinical trials do not involve treatment and drugs.
-
DrugBank. DrugBank11 (https://www.drugbank.com/) is a comprehensive, freely accessible online database that provides detailed information about drugs and their biological targets. We extract the drug molecular structures and pharmaceutical properties from DrugBank, which are essential to drug’s safety in human bodies and efficacy in treating certain diseases.
-
TrialTrove. TrialTrove (https://pharmaintelligence.informa.com/products-and-services/data-and-analysis/trialtrove) is a comprehensive database and intelligence platform designed to provide detailed information and analysis on clinical trials across the pharmaceutical and biotechnology industries. TrialTrove serves as a critical resource for professionals involved in clinical development, competitive intelligence, and market analysis. We obtain the trial outcomes of some trials from the released/public subset of the TrialTrove database23.
-
ICD-10. ICD-10-CM (International Classification of Diseases, 10th Revision, Clinical Modification) is a medical coding system for classifying diagnoses and reasons for visits in U.S. healthcare settings. Diseases are extracted from https://clinicaltrials.gov/ and linked to ICD-10 codes and disease description using Clinical Table Search Service API (clinicaltables.nlm.nih.gov) and then to CCS codes via hcup-us.ahrq.gov/toolssoftware/ccs10/ccs10.jsp.
We collect the AI-ready input and output information by (1) extracting treatment names (e.g., drug names) from ClinicalTrials.gov and linking them to its molecule structure (SMILES strings and the molecular graph structures) using the DrugBank Database; (2) extracting disease data from ClinicalTrials.gov and linking them to ICD-10 (International Classification of Diseases, Tenth Revision) codes and disease description using clinicaltables.nlm.nih.gov and then to CCS codes via hcup-us.ahrq.gov/toolssoftware/ccs10/ccs10.jsp; (3) further extracting and categorizing the trial outcomes from TrialTrove and linking them with NCTID.
Dataset Curation and Feature Organization
We apply a series of selection filters to ensure the selected trials have high-quality. There are hundreds of multi-modal features in ClinicalTrials.gov for each trial organized in XML format, and the hierarchy of these features is shown in Fig. S1. We only leverage the features that are available before trials start and remove the remaining features. Different tasks rely on different subsets of features. Based on clinical trial knowledge, we manually select the appropriate features for various tasks. In addition, we also remove features whose values are identical or all null across different trials. Following are the additional selection criteria for each task.
-
Trial duration prediction: We only consider the trials whose start and completion dates are available. We only consider the trials with realistic completion dates and remove the cases with only anticipated completion dates provided. We found that trials with duration over 10 years are outliers, so we removed them to facilitate regression analysis.
-
Patient dropout prediction: The results are available at ClinicalTrials.gov and the number of dropout and total enrolled patients are reported.
-
Adverse event prediction: The results are available at ClinicalTrials.gov and the serious adverse events are reported.
-
Mortality event prediction: The results are available at ClinicalTrials.gov and mortality event is reported.
-
Trial approval prediction: The results and trial outcome information are available at either ClinicalTrials.gov or the released subset of TrialTrove23.
-
Trial failure reason identification: We incorporate those trials whose results and outcome information are available at ClinicalTrials.gov and can be categorized into four categories (three failure reasons or success) mentioned above.
-
Eligibility criteria design: To ensure the high quality of the selected eligibility criteria, we only incorporate completed trials, indicating successful patient recruitment and reasonable criteria design, and remove the others.
-
Drug dose finding: We incorporate trials whose drug dosage information is available on ClinicalTrials.gov. Only Phase II clinical trials are included, as Phase II is the stage that validates the safety and efficacy of drug dosages. Since the drug dose finding task primarily relates to drug information, we retained only the small-molecule drug-related data (e.g., MeSH) and sourced SMILES from DrugBank. We encourage AI experts to utilize external knowledge from sources such as PubMed and DrugBank for advanced AI model development24,25.
Apart from flattening the XML nodes and attributes into tabular features, we also specially pre-process several features to be more deep learning approach-ready formats: We transform the information recorded in the XML node named “ipd_info_type” into multiple tabular features. The “ipd_info_type” feature specified the provided document types provided such as “Study Protocol”, ‘Statistical Analysis Plan (SAP)”, “Informed Consent Form (ICF)”, and “Clinical Study Report (CSR)”. In one clinical trial, several types of documents may be provided. Thus, we conveyed such information into multiple binary features, where each document type is represented in a binary categorical feature. The columns are named as “ipd_info_type-Analytic Code”, “ipd_info_type-Clinical Study Report (CSR)”, “ipd_info_type-Informed Consent Form (ICF)”, “ipd_info_type-Statistical Analysis Plan (SAP)”, and “ipd_info_type-Study Protocol”, respectively. If a document type appears in the data, the corresponding column value is 1; otherwise, it is 0. Similar strategies were applied on other nodes presenting discrete values, like “study_design_info/masking”, “arm_group/arm_group_type”, and “intervention/intervention_type”.
Data Annotation
Data annotation (a.k.a. labeling data) is a fundamental step when curating a dataset. Labels of all the datasets can be inferred from various data sources. For some tasks, such as drug dose finding, trial approval prediction, and trial failure reason identification, we use external tools such as GPT to obtain the label from the raw text.
• Trial duration prediction: The duration of a clinical trial refers to the number of years the trial lasts, i.e., the difference between the start and complete date. It is a continuous numerical value. For some trials, the start and completion date are available in ClinicalTrials.gov. We can use this information to calculate the trial duration.
• Patient dropout prediction: Some clinical trials on ClinicalTrials.gov present the number of dropout patients and the number of enrolled patients. We compute the patient dropout rate by dividing the number of dropout patients by the number of enrolled patients. The resulting dropout rate is a percentage.
• Adverse event prediction: ClinicalTrials.gov presents the results of some trials. Adverse events are reported for some of these trials.
• Mortality event prediction: The results of clinical trials presented on ClinicalTrials.gov may include mortality events. We binarize the mortality event as the prediction target indicating whether a mortality event occurred, and remove all other trials that lack mortality event information.
• Trial approval prediction: The annotations come from two sources. First, the HINT paper23,26,27,28 builds a benchmark dataset for trial approval prediction, with approval labels sourced from TrialTrove. Additionally, ClinicalTrials.gov provides termination reasons for some trials, such as poor enrollment or lack of efficacy, included in the “why stopped” node in the XML files. We incorporate these trials, along with termination reasons indicating failed approval, into the dataset as negative samples.
• Trial failure reason identification: For some of the terminated trials, ClinicalTrials.gov provides a “why stopped” tag that uses natural language to describe the failure reason. We use OpenAI ChatGPT API (https://openai.com/index/openai-api/) to automatically convert into four categories of failure reason, including (1) poor enrollment; (2) drug safety issue; (3) lack of efficacy (in treating the target disease); (4) others (e.g., lack of funding, strategic decision by sponsor). Since the last failure reason ((4) others) is usually not predictable, we perform 4-category classification ((1) success; (2) poor enrollment; (3) drug safety issue (4) lack of efficacy). In using ChatGPT, the prompt and instruction are shown below, and we required ChatGPT to complete the “reasons” part: 
We input “why stopped” contexts of 10 clinical trials into ChatGPT in each iteration. We also use the passed trials from the released subset of TrialTrove, following23.
• Eligibility criteria design: For some trials, the eligibility criteria are organized in a textual format and are available on ClinicalTrials.gov. We considered the inclusion/exclusion eligibility criteria of trials marked as “completed” as the ground truth.
• Drug dose finding: One aim of phase-II clinical trials is to determine the dosage of the drug. ClinicalTrials.gov presents the drug dosage information of some trials in natural language. We use OpenAI ChatGPT API (https://openai.com/index/openai-api/) to extract the label from natural language, the prompt is shown below. 
We categorize these doses into four classes: (1): dose < 1 mg/kg; (2) 1 mg/kg < dose < 10 mg/kg; (3) 10 mg/kg < dose < 100 mg/kg; (4) dose > 100 mg/kg. For dosages expressed in units such as mg per person or mg/hour, we assume an individual weight of 60 kg and convert using 24 hours per day to keep the units consistent .
Data Partitioning
We adopt random partitioning for dataset splitting. For classification tasks, stratified sampling is applied to preserve class distribution across training and test sets; for regression tasks, random splitting is used. The default split ratio is 80/20.
To promote robust model development, we encourage users to explore alternative, task-relevant splitting strategies. For instance, a temporal split—training on earlier trials and testing on later ones—can emulate real-world deployment scenarios. Standard approaches such as five-fold cross-validation may also be employed to assess model robustness. Additionally, location-based splitting can be used to evaluate geographic generalizability, which is particularly relevant for tasks such as predicting patient dropout or engagement.
Ethics Statement
The development and dissemination of the TrialBench dataset adhere to stringent ethical standards to ensure the protection of patient privacy, the integrity of the data, and the responsible use of the information. The source of the data is clearly documented, and proper attribution is given to ClinicalTrials.gov and other databases such as DrugBank11 and TrialTrove. This transparency ensures that users of the TrialBench dataset understand the origin of the data and the context in which it was collected.
Data Records
Our curated datasets, collectively referred to as TrialBench, are publicly available at https://huyjj.github.io/Trialbench/, accompanied by detailed documentation and illustrative diagrams to facilitate comprehension. For practical use, the data can be accessed and manipulated through our Python and R packages, with installation instructions available on the project website.
Data Overview
Clinical trial records are originally organized in an XML hierarchy format, we selected relevant features based on the challenges of each task and re-organized them into a tabular data format. Notably, in addition to categorical and numerical tabular features, some of these features may include free text, graph data, and other complex types.
Here, we review some essential features in our datasets. Notably, some trials have missing features; for example, certain incomplete trials lack a completed date and outcome.
• Trial questions. A clinical trial aims to answer the question: Is the treatment effective in treating the target diseases for patients? First, the treatment must be safe for the human body. Second, the new drug candidate should be better than the current standard treatment.
• National Clinical Trial number (NCT ID) is the identifier of the clinical trial. It consists of 11 characters and begins with NCT, e.g., NCT02929095. NCT ID is assigned based on the temporal order of registration date and starts from NCT00000000.
• Study type. Clinical trials can be categorized into interventional and observational. Interventional clinical trials involve drugs, medical devices, or surgery as treatment. In contrast, observational trials do not assign participants to a treatment or other intervention. Instead, the researchers observe participants or measure certain outcomes to determine clinical outcomes.
• Phase. Phase I tests the toxicity and side effects of the drug; phase II determines the efficacy of the drug (i.e. if the drug works); phase III focuses on the effectiveness of the drug (i.e., whether the drug is better than the current standard practice). When the trial passes phase III, it can be submitted to the FDA for approval. In many cases, even after approval, we still need to further monitor the drugs’ effectiveness and safety. Sometimes a phase IV trial will be conducted to assess the drug’s effectiveness and safety. Table 4 demonstrate the differences between phases I, II, III, and IV.
• Eligibility criteria describe the patient recruitment requirements in unstructured natural language. Eligibility criteria comprise multiple inclusion and exclusion criteria, which specify what is desired and undesired when recruiting patients. Each individual criterion is usually a natural language sentence. For example, in the clinical trial entitled “Efficacy and Safety Study of MP-513 in Combination With Thiazolidinedione in Patients With Type 2 Diabetes”(https://clinicaltrials.gov/ct2/show/NCT01026194), which is a phase III trial, the inclusion criteria contain:

The exclusion criteria contain:

• Disease (also known as condition, or indication) describes the diseases that the drug is intended to treat. It is in unstructured natural language. For example, NCT00428389 studies the safety of switching from Donepezil to Rivastigmine patch in patients with probable Alzheimer’s Disease, where Alzheimer’s disease is the disease that the trial wants to treat. Sometimes, a single trial may target multiple diseases or patients with co-morbidities.
• Disease code. The disease is usually described by natural language, and it is hard to reveal the relationship between different diseases29,30,31. To address this issue, we map disease names to disease codes and leverage the disease hierarchy for machine learning modeling. For example, several ICD-10 codes correspond to Alzheimer’s disease, including “G30.0” (Alzheimer’s disease with early onset), “G30.1” (Alzheimer’s disease with late onset), “G30.8” (Other Alzheimer’s disease), “G30.9” (Alzheimer’s disease, unspecified)24,32.
• Title of the clinical trial is usually in unstructured natural language.
• Summary of the clinical trial is also in terms of unstructured natural language, which consists of 2-5 sentences that describe the tested treatment, target disease to treat, and the main objective of the clinical trial.
• Study type. There are mainly two study types: interventional and observational. Interventional trials assess an intervention/treatment, which can be drugs, medical devices, surgery, activity (exercise), procedure, etc. In contrast, observational trials do not involve an intervention or treatment; instead, in observational trials, patients take normal treatment, researchers observe/track patients’ health records and analyze the results. We restrict our attention to the subset of interventional trials using drug candidates as the interventions.
• Drug (also known as intervention or treatment). In the trial document, the drug names are shown. We also know the category of the drug, i.e., whether it belongs to small-molecule drug or biologics. The treatment usually involves one or multiple drug molecules. We can also map the drug candidate to its molecule structure, such as its SMILES string (The simplified molecular-input line-entry system (SMILES) is a specification in the form of a line notation for describing the structure of chemical species using short ASCII strings).
• Trial site. One trial is usually conducted in multiple trial sites so that scientists can recruit sufficient patients. Scientists also hope to reduce the bias of patient groups and enhance their diversity, so the geographic location of the trial sites is also considered.
• Patient. The trial runner need to recruit eligible patient volunteers based on their electronic health records (EHR) in the trial sites to conduct the trial. The requirement of recruiting patients is provided in the eligibility criteria.
• Electronic Health Record (EHR). An electronic health record (EHR) is the longitudinal digital record of patients and contains patients’ medical histories. The growing volume and availability of Electronic Health Record (EHR) data have sparked an interest in using machine learning methods for supporting drug development. For example, machine learning approaches such as33,34 have been proposed to map patient EHR data to clinical trial eligibility criteria. EHR data comprises medical records of N different patients. The medical record of each patient is longitudinal data.
• Start date is the registration date of the clinical trial. NCTID is assigned based on the order of start date.
• Completion date refers to the date when the clinical trial is complete. Incomplete clinical trials have the expected completion dates.
• Sponsors of the clinical trial can be pharmaceutical companies or research institutes. For example, the trial entitled “PF-06863135 As Single Agent And In Combination With Immunomodulatory Agents In Relapse/Refractory Multiple Myeloma”(https://clinicaltrials.gov/ct2/show/NCT03269136) is supported by Pfizer; the trial entitled “Five, Plus Nuts and Beans for Kidneys” (https://clinicaltrials.gov/ct2/show/NCT03299816) is supported by Johns Hopkins University. Some trials may contain multiple sponsors. Table 5 lists the top 20 sponsors that conduct the most interventional clinical trials.
• Outcome. Generally, the trial outcomes are usually complex, involving many statistics and analyses. In some tasks, such as clinical trial outcome prediction, the outcome can be abstracted into binary labels, e.g., whether the tested drug passed a particular phase.
• Failure reason. Clinical trials suffer from high failure rates due to multiple reasons, including business decisions (e.g., lack of funding, company strategy shift), poor enrollment, drug safety issues (e.g., adverse effects), and lack of efficacy.
Summarization of Multi-Modal Features
Clinical trials involve diverse modalities of data, as shown in the following.
Categorical Features
Categorical features typically describe some qualitative attributes. For example, there are mainly two study types: interventional and observational. The intervention type can be a small-molecule drug, biologics, or surgery, etc. Clinical trial sponsors can be pharmaceutical companies or research institutes, e.g., Johns Hopkins University, or Pfizer.
Numerical Features
Numerical features, such as the minimum/maximum age of recruited patients and the number of real/expected recruited patients, represent quantitative data, are also common in clinical trials. Numerical features, along with categorical features, are two important types of tabular features35,36.
Text Features
In clinical trials, there are many text features that contain rich information for AI modeling. For example, eligibility criteria describe the patient recruitment requirements in unstructured natural language; each clinical trial contains a summary, which consists of 2-5 natural language sentences that describe the tested treatment, the target disease to treat, and the main objective of the clinical trial. To process such datasets, we treat the text data as sequences of tokens (e.g., words). How to extract useful information from unstructured text has been extensively studied with several well-known deep neural network architectures, such as recurrent neural network (RNN)37, convolutional neural network (CNN), and transformer architecture38.
Drug Molecule
The most expressive and intuitive data representation of a drug molecule is the 2D molecular graph39, where each node corresponds to an atom in the molecule while an edge corresponds to a chemical bond. The molecular graph mainly contains two essential components: node identities and node interconnectivity. The nodes’ identities include atom types, e.g., carbon, oxygen, nitrogen, etc. The nodes’ connectivity can be represented as an adjacency matrix, where the (i,j)-th element denotes the connectivity between i-th and j-th nodes.
MeSH Terms
The Medical Subject Headings (MeSH) comprehensively index, catalog, and search biomedical and health-related information. It consists of sets of terms in a hierarchical structure that enables more precise and efficient retrieval of information. Unlike ICD-10, which primarily classifies diseases and medical conditions, MeSH is also used to index and retrieve information on broader health-related topics such as anatomy, drugs, and diseases.
Disease Code
There are several standardized disease coding systems that healthcare providers use for the electronic exchange of clinical health information, including the International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM), The International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM), and Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT)40. These coding systems contain disease concepts organized into hierarchies. We take the ICD-10-CM code as an example. ICD-10-CM is a seven-character, alphanumeric code. Each code begins with a letter, and two numbers follow that letter. The first three characters of ICD-10-CM are the “category”. The category describes the general type of injury or disease. A decimal point and the subcategory follow the category. For example, the code “G44” represents “Other headache syndromes”; the code “G44.31” represents “Acute post-traumatic headache”; the code “G44.311” represents “Acute post-traumatic headache, intractable”. G44.311 has two ancestors: G44 and G44.31, where an ancestor represents a higher-level category of the current code. The description of all the ICD-10-CM codes is available at https://www.icd10data.com/ICD10CM/Codes. We also illustrate the hierarchy in Fig. 3.
Technical Validation
To show the processed datasets are AI-ready and of reasonable quality, we evaluate the performance of these datasets on mainstream AI algorithms. We leverage a multi-modal deep neural network to represent the multi-modal features and concatenate all these representations to make the prediction. In this section, we first discuss the multi-modal deep learning method, then describe the experimental setup, and finally present the experimental results.
Multi-modal Deep Neural Networks
For all classification and regression tasks, we apply various deep neural networks to represent multimodal features. Each representation is an embedding vector with continuous values. Then, we concatenate these representations, feed them into multiple-layer perceptron (MLP), and make the prediction. For the eligibility criteria design task, we use OpenAI ChatGPT API (https://openai.com/index/openai-api/) with the prompt to produce eligibility criteria.
Categorical and Numerical Tabular Features
Recently, numerous tabular data processing models35,36,41 have been proposed for numerical and categorical feature processing. Among them, DANets42 stand out due to its key component’s modularity and ability to achieve competitive performance without hyperparameter tuning. The key component, the basic block module, supports flexible stacking, making DANets suitable as a submodule for processing numerical and categorical features. After preprocessing (e.g., normalization), three lightweight basic blocks are sequentially stacked to hierarchically select, extract, and merge features from input categorical and numerical features, ultimately yielding a 50-dimensional embedding.
Disease Code
Graph-based Attention Model (GRAM) is an attention-based neural network model that leverages the hierarchical information inherent to disease codes (medical ontologies)43. Specifically, each disease code is assigned a basic embedding, e.g., the disease code di has basic embedding, denoted \({{\bf{e}}}_{i}\in {{\mathbb{R}}}^{d}\). Then, to impute the hierarchical dependencies, the embedding of current disease di (denoted hi) is represented as a weighted average of the basic embeddings (\({\bf{e}}\in {{\mathbb{R}}}^{d}\)) of itself and its ancestors, the weight is evaluated by the attention model. It is formally defined as
where αji ∈ (0, 1) represents the attention weight and is defined as
where the attention model ϕ(⋅) is an MLP with a single hidden layer, the input is the concatenation of the basic embedding, the output is a scalar, ei serves as the query while all the ancestors embeddings \(\left\{{{\bf{e}}}_{j}\right\}\) serve as the keys. Ancestors (i) represents the set of all the ancestors of the disease code di. The GRAM model is illustrated in Fig. 4.
MeSH Terms
Similar to modern word embeddings that represent word semantics, Medical Subject Headings (MeSH) codes from the MeSH thesaurus can also be represented using embedding approaches. MeSH-Embedding44 has pretrained a MeSH embedding layer using the node2vec algorithm45 with default parameters. For MeSH terms that have not been included in pretraining the MeSH embedding layer44, we employ a new parametric embedding layer learned from scratch.
Text Features
Bidirectional Encoder Representations from Transformers (BERT)46 is a powerful pretraining technique that has its roots in the Transformer architecture and was specifically designed for natural language processing (NLP) tasks. In recent years, it has been widely applied to drug discovery and has proven to be effective in modeling text data. BERT is constructed by stacking multiple layers of Transformer blocks. The output of each layer is used as the input to the subsequent layer, thus allowing the model to learn increasingly complex representations of the input data. This technique results in a deep, bidirectional architecture that is capable of capturing contextual information from both the past and future tokens in a sequence. The key advantage of using BERT for this task is that it enables the model to leverage the knowledge learned from the massive unlabeled data to better understand the relationships between the sequences and their corresponding properties. This allows the model to make more accurate predictions compared to training the model from scratch using only the limited labeled data available for the specific task. In this paper, we use Bio-BERT47, a variant of BERT that is pretrained in biomedical literature.
Drug Molecule
Drug Molecule is essentially 2D planar graph. Graph neural network (GNN) is a neural network architecture that takes graph-structured data as input, transmits the information between the connected edges and nodes to capture the interaction between them, and learns a vector representation of graph nodes and the entire graph48. Message Passing Neural Network (MPNN)49 is a popular variant of GNN, which updates the information of edges in a graph. First, on the node level, each node v has a feature vector denoted ev. For example, node v in a molecular graph G is an atom, ev includes the atom type, valence, and other atomic properties. ev can be a one-hot vector indicating the category of the node v. On the edge level, euv is the feature vector for edge (u, v). \({\mathcal{N}}(u)\) represents the set of all the neighbor nodes of the node u. At the l-th layer, \({{\bf{m}}}_{uv}^{(l)}\) and \({{\bf{m}}}_{vu}^{(l)}\) are the directional edge embeddings representing the message from node u to node v and vice versa. They are iteratively updated as
where ⊕ denotes the concatenation of two vectors; f1(⋅) is a multiple layer perceptron (MLP), \({{\bf{m}}}_{uv}^{(l)}\) is the message vector from node u to node v at the l-th iteration, whose initialization is all-0 vector, i.e., \({{\bf{m}}}_{uv}^{(0)}={\bf{0}}\), following the rule of thumb50. After L steps of iteration (L is the depth), another multiple layer perceptron (MLP) f2(⋅) is used to aggregate these messages. Each node has an embedding vector as
We are interested in graph-level representation hG, we can further use the readout function (e.g., average) to aggregate all the node embeddings.
Representation Fusion
After obtaining the representations of multi-modal data, we concatenate these representations, feed the concatenated vector into a multiple-layer perceptron (MLP), and make the prediction. For binary classification tasks (e.g., trial approval prediction), we use the sigmoid function as the activation function in the output layer to yield predicted probability; for multi-category classification tasks (e.g., trial failure reason identification), we use softmax as the activation function in the output layer to produce probability distribution over all the categories; for regression tasks (e.g., trial duration prediction), we do not use activation function in the output layer to produce continuous-valued prediction. We use cross-entropy criterion as the loss function for classification tasks and mean-square error (MSE) as the loss function for regression tasks.
Experimental Setup
Implementation Details
All the code is implemented in Python 3.8. All deep learning models are implemented in PyTorch, and we use GPT 4.0 for data annotation and generation tasks. The embedding size of all the representations is set to 100. We use Adam51,52 as the numerical optimizer to minimize the loss function with an initial learning rate at 1e − 3 and zero weight decay. The batch size is set to 64. The maximal training epochs is set to 20.
Evaluation Metrics
For classification tasks, we assess the model performance using accuracy, PR-AUC (the area under the Precision-Recall curve), F1 score (the harmonic mean of precision and recall), Precision, Recall, Specificity, and ROC-AUC (the Area Under the Receiver Operating Characteristic Curve). For regression tasks, we use RMSE (Root Mean Squared Error), MAE (Mean Absolute Error), Concordance Index, and Pearson Correlation as metrics. For generation tasks (eligibility criteria design), we design some semantic metrics to measure the alignment between real and designed criteria, including text embeddings’ cosine similarity, informativeness, and redundancy, detailed in the Supplementary Information.
Validation Results
In this section, we demonstrate the experimental results of multi-modal deep learning methods on all the curated tasks and datasets in Table 6. We find that the direct use of a multimodal deep learning method leads to decent performance in most of the curated tasks. Specifically, for 14 binary classification datasets (across patient dropout prediction, adverse event prediction, mortality event prediction, and trial approval prediction), the multimodal deep learning method achieves at least 0.7 F1 scores in 11 datasets. On regression and generation tasks, the simple multi-modal deep learning method also achieves decent performance. These results validate the AI-readiness and high quality of the curated datasets.
Usage Note
This paper extracts various properties of clinical trials and integrates them with multiple data sources. These properties are essential for analyzing and predicting different aspects of clinical trial performance and outcomes. The properties extracted include:
-
Trial duration: The length of time a clinical trial lasts, from its start date to its completion date. This helps in understanding the efficiency and planning required for trials.
-
Patient dropout: The premature withdrawal of participants from a trial. High dropout rates may compromise statistical power and introduce bias, thereby affecting the integrity of the study results.
-
Adverse event: Instances of significant negative health effects observed during the trial, which are crucial for evaluating the safety profile of the treatment being tested.
-
Mortality event: Occurrences of participant death during the trial. This metric is critical for evaluating the potential risks associated with the treatment.
-
Trial approval outcome: Whether a drug can pass a certain phase of the clinical trial, which is a binary outcome indicating success or failure.
-
Trial failure reason: The identification of reasons why a clinical trial may fail, such as poor enrollment, safety issues, or lack of efficacy. This helps in improving the design of future trials.
-
Eligibility criteria design: The inclusion and exclusion criteria for participants are essential for ensuring that the right population is targeted for the trial.
-
Drug dosage: Estimating the appropriate dosage of drugs being tested to ensure safety and efficacy.
These properties and the datasets provided in this study enable researchers and AI practitioners to apply advanced machine learning models to predict and optimize various aspects of clinical trials. The datasets include multi-modal data, such as drug molecules, disease codes, textual descriptions, and categorical/numerical features, making them versatile for different predictive tasks. By leveraging these datasets, researchers can improve clinical trial design, enhance patient safety, optimize resource allocation, and ultimately accelerate the development of new medical treatments.
Intended Users
TrialBench is intended for healthcare, biomedical, and AI researchers and data scientists who want to apply AI algorithms and innovate novel methods to tackle problems formulated in TrialBench datasets and tasks.
Demonstration of Python and R Package Usage
Users can conveniently download datasets via the trialbench package, available in both Python and R. In Python, the data can be loaded either as a PyTorch-compatible DataLoader format or as a DataFrame format using the pandas library, facilitating downstream model development. In R, the data is provided in the standard data.frame structure for seamless integration with typical R workflows. Examples are illustrated as in Code 1.
Code 1.
TrialBench data downloading using python or R packages.
Computing Resources
We use a server with an NVIDIA GeForce RTX 3090 GPU, Intel(R) Xeon(R) CPU with 50GB RAM for all empirical experiments in this manuscript.
Limitations
Artificial intelligence for clinical trial is a vast and fast-growing field, and there are important tasks and datasets yet to be included in TrialBench. However, TrialBench is an ongoing effort and we strive to continuously include more datasets and tasks in the future.
Licensing
Most of the data features come from ClinicalTrials.gov, which is a service of the U.S. National Institutes of Health, provides access to information on publicly and privately supported clinical studies. The data available on ClinicalTrials.gov is generally free for use. Some TrialBench tasks involve data in DrugBank, which is available for free to academic institutions and non-profit organizations for research and educational purposes. The subset of TrialTrove is released by Fu’s study23 and is publicly available for Non-Commercial Use.
Code availability
The codes to reproduce results in Table 6 can be retrieved from the Github repository at https://github.com/ML2Health/ML2ClinicalTrials/tree/main/Trialbench.
References
Piantadosi, S.Clinical trials: a methodologic perspective (John Wiley & Sons, 2024).
Hackshaw, A. A concise guide to clinical trials (John Wiley & Sons, 2024).
Eichler, H.-G. & Sweeney, F. The evolution of clinical trials: Can we address the challenges of the future? Clin. Trials 15, 27–32 (2018).
Sun, D., Gao, W., Hu, H. & Zhou, S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B 12, 3049–3062 (2022).
Martin, L., Hutchens, M., Hawkins, C. & Radnov, A. How much do clinical trials cost. Nat. Rev. Drug Discov. 16, 381–382 (2017).
Lipkova, J. et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell 40, 1095–1110 (2022).
Askin, S., Burkhalter, D., Calado, G. & El Dakrouni, S. Artificial intelligence applied to clinical trials: opportunities and challenges. Health Technol. 13, 203–213 (2023).
Acosta, J. N., Falcone, G. J., Rajpurkar, P. & Topol, E. J. Multimodal biomedical ai. Nat. Med. 28, 1773–1784 (2022).
Huang, K. et al. Therapeutics data commons: machine learning datasets and tasks for therapeutics. NeurIPS Track Datasets and Benchmarks (2021).
Huang, K. et al. Artificial intelligence foundation for therapeutic science. Nat. Chem. Biol. 1–4 (2022).
Wishart, D. S. et al. Drugbank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018).
Glick, H. A., Doshi, J. A., Sonnad, S. S. & Polsky, D. Economic evaluation in clinical trials (OUP Oxford, 2014).
Alexander, W. The uphill path to successful clinical trials: keeping patients enrolled. Pharmacy and Therapeutics 38, 225 (2013).
Singh, S. & Loke, Y. K. Drug safety assessment in clinical trials: methodological challenges and opportunities. Trials 13, 1–8 (2012).
Van Gerven, J. & Bonelli, M. Commentary on the ema guideline on strategies to identify and mitigate risks for first-in-human and early clinical trials with investigational medicinal products. Br. J. Clin. Pharmacol. 84, 1401 (2018).
Silverman, H. Ethical issues during the conduct of clinical trials. Proc. Am. Thorac. Soc. 4, 180–184 (2007).
Friedman, L. M., Furberg, C. D., DeMets, D. L., Reboussin, D. M. & Granger, C. B. Fundamentals of clinical trials (Springer, 2015).
Kobak, K. A., Kane, J. M., Thase, M. E. & Nierenberg, A. A. Why do clinical trials fail?: the problem of measurement error in clinical trials: time to test new paradigms? J. Clin. Psychopharmacol. 27, 1–5 (2007).
Chow, S.-C., Shao, J., Wang, H. & Lokhnygina, Y. Sample size calculations in clinical research (chapman and hall/CRC, 2017).
Peters-Lawrence, M. H. et al. Clinical trial implementation and recruitment: lessons learned from the early closure of a randomized clinical trial. Contemp. Clin. Trials 33, 291–297 (2012).
Ting, N. Dose finding in drug development (Springer Science & Business Media, 2006).
Chang, Y.-T. et al. Integrated identification of disease specific pathways using multi-omics data. Cold Spring Harb. Lab. 666065 (2019).
Fu, T., Huang, K., Xiao, C., Glass, L. M. & Sun, J. HINT: Hierarchical interaction network for clinical-trial-outcome predictions. Patterns 3, 100445 (2022).
Chen, L. et al. Data-driven detection of subtype-specific differentially expressed genes. Sci. Rep. 11, 332 (2021).
Lu, Y., Sato, K. & Wang, J. Deep learning based multi-label image classification of protest activities. arXiv preprint arXiv:2301.04212 (2023).
Chen, T., Hao, N., Lu, Y. & Van Rechem, C. Uncertainty quantification on clinical trial outcome prediction. Health Data Sci. (2024).
Chen, T., Hao, N., Van Rechem, C., Chen, J. & Fu, T. Uncertainty quantification and interpretability for clinical trial approval prediction. Health Data Sci. 4, 0126 (2024).
Wang, Y. et al. TWIN-GPT: Digital Twins for Clinical Trials via Large Language Model. ACM Trans. Multimedia Comput. Commun. Appl. (2024).
Lu, Y. Multi-omics Data Integration for Identifying Disease Specific Biological Pathways. Ph.D. thesis, Virginia Tech (2018).
Wu, C.-T. et al. Cosbin: cosine score-based iterative normalization of biologically diverse samples. Bioinformatics Adv. 2, vbac076 (2022).
Fu, Y. et al. Ddn3. 0: Determining significant rewiring of biological network structure with differential dependency networks. Bioinformatics btae376 (2024).
Lu, Y. et al. COT: an efficient and accurate method for detecting marker genes among many subtypes. Bioinformatics Adv. 2, vbac037 (2022).
Zhang, X., Xiao, C., Glass, L. M. & Sun, J. Deepenroll: Patient-trial matching with deep embedding and entailment prediction. In Proceedings of The Web Conference 2020, 1029–1037 (2020).
Gao, J., Xiao, C., Glass, L. M. & Sun, J. COMPOSE: Cross-modal pseudo-siamese network for patient trial matching. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, 803–812 (2020).
Chen, J. et al. Excelformer: Can a dnn be a sure bet for tabular prediction? In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining (2024).
Gorishniy, Y., Rubachev, I., Khrulkov, V. & Babenko, A. Revisiting deep learning models for tabular data. Adv. Neural Inf. Process. Syst. 34, 18932–18943 (2021).
Hochreiter, S. & Schmidhuber, J. Lstm can solve hard long time lag problems. Adv. Neural Inf. Process. Syst. 9 (1996).
Vaswani, A. et al. Attention is all you need. In Advances in neural information processing systems, 5998–6008 (2017).
Coley, C. W., Barzilay, R., Green, W. H., Jaakkola, T. S. & Jensen, K. F. Convolutional embedding of attributed molecular graphs for physical property prediction. J. Chem. Inf. Model. 57, 1757–1772 (2017).
Anker, S. D., Morley, J. E. & von Haehling, S. Welcome to the ICD-10 code for sarcopenia. J. Cachexia Sarcopenia Muscle 7, 512–514 (2016).
Chen, J., Liao, K., Fang, Y., Chen, D. & Wu, J. Tabcaps: A capsule neural network for tabular data classification with bow routing. In The Eleventh International Conference on Learning Representations (2022).
Chen, J., Liao, K., Wan, Y., Chen, D. Z. & Wu, J. DANETs: Deep abstract networks for tabular data classification and regression. In Proceedings of the AAAI Conference on Artificial Intelligence, 3930–3938 (2022).
Choi, E., Bahadori, M. T., Song, L., Stewart, W. F. & Sun, J. GRAM: graph-based attention model for healthcare representation learning. In Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, 787–795 (2017).
Helboukkouri, I. Mesh embeddings. https://github.com/helboukkouri/mesh-embeddings (ongoing). [Accessed: 2024-06-02].
Grover, A. & Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD international conference on Knowledge discovery and data mining, 855–864 (2016).
Devlin, J., Chang, M., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In NAACL-HLT, 4171–4186 (Association for Computational Linguistics, 2019).
Lee, J. et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36, 1234–1240 (2020).
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. Int. Conf. Learn. Represent. (ICLR) (2016).
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O. & Dahl, G. E. Neural message passing for quantum chemistry. In International conference on machine learning, 1263–1272 (PMLR, 2017).
Fu, T., Xiao, C. & Sun, J. CORE: Automatic molecule optimization using copy & refine strategy. In Proceedings of the AAAI Conference on Artificial Intelligence, 638–645 (2020).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. Int. Conf. Learn. Represent. (ICLR) (2014).
Chen, J. et al. TrialBench: Multi-modal AI-ready datasets for clinical trial prediction, https://huyjj.github.io/Trialbench/ (2025).
Acknowledgements
Tianfan Fu is supported by Nanjing University International Collaboration Initiative. Jintai Chen is supported by internal funding from the Hong Kong University of Science and Technology (Guangzhou).
Author information
Authors and Affiliations
Contributions
The project was designed by J. Chen, J. Sun, L. Glass, M. Zitnik, and T. Fu. J. Chen, Y. Hu, M. Cai and Y. Wang curated the datasets. Y. Hu, Y. Lu, Y. Wang, and T. Fu developed and validated the model. J. Chen, X. Cao, K. Huang, and T. Fu drafted the paper, while J. Chen, Y. Lu, M. Lin, H. Xu, J. Wu, K. Huang, and T. Fu reviewed and proofread the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, J., Hu, Y., Cai, M. et al. TrialBench: Multi-Modal AI-Ready Datasets for Clinical Trial Prediction. Sci Data 12, 1564 (2025). https://doi.org/10.1038/s41597-025-05680-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05680-8