Abstract
Multimodal datasets offer valuable support for developing Clinical Decision Support Systems (CDSS), which leverage predictive models to enhance clinicians’ decision-making. In this observational study, we present a dataset of suspected Coronary Artery Disease (CAD) patients - called MultiD4CAD - comprising imaging and clinical data. The imaging data obtained from Coronary Computed Tomography Angiography (CCTA) includes epicardial (EAT) and pericoronary (PAT) adipose tissue segmentations. These metabolically active fat tissues play a key role in cardiovascular diseases. In addition, clinical data include a set of biomarkers recognized as CAD risk factors. The validated EAT and PAT segmentations make the dataset suitable for training predictive models based on radiomics and deep learning architectures. The inclusion of CAD disease labels allows for its application in supervised learning algorithms to predict CAD outcomes. MultiD4CAD has revealed important correlations between imaging features, clinical biomarkers, and CAD status. The article concludes by discussing some challenges, such as classification, segmentation, radiomics, and deep training tasks, that can be investigated and validated using the proposed dataset.
Similar content being viewed by others
Background & Summary
Coronary Artery Disease (CAD) is one of the main causes of death worldwide. This disease can lead to angina pectoris, myocardial infarction, heart failure, and sudden death1. Fat tissues surrounding the heart demonstrate different fundamental roles. Several studies have shown that Epicardial Adipose Tissue (EAT) — involved in coronary vessel mechanical support, energy reserve, and thermoregulatory function — is also implicated in the development of CAD and plays an important role in cardiovascular events prediction2. Moreover, the Pericoronary adipose tissue (PAT) — a part of the EAT located around the coronary arteries — plays an active role in inflammatory and metabolic regulation, directly influencing cardiovascular health. Understanding these mechanisms is essential for developing more effective prevention and treatment strategies for CAD3. Several studies prove that PAT represents a biomarker in CAD assessment4 and in different clinical settings5.
In this scenario, the detection of high-risk atherosclerotic plaque biomarkers (i.e., low attenuation, positive remodeling, patchy calcification, and napkin-ring sign) in Coronary Computed Tomography Angiography (CCTA) allows highly specific labeling of patients at high risk for major adverse cardiac events. It was shown how these biomarkers are predictive of ischemia, even in nonobstructive lesions6. With more details, recent studies have focused on CT attenuation of EAT and PAT as an indirect marker of coronary atherosclerosis and plaque inflammation7. Inflammation is a crucial component of atherosclerosis and a consistent pathological feature of unstable atherosclerotic plaques. Increased CT attenuation in adipose tissue adjacent to an atherosclerotic plaque is a marker of inflammation8,9.
Considering the discussed substantial role of EAT and PAT in cardiovascular diseases, this retrospective observational study aims to define and propose a dataset composed of EAT and PAT Regions of Interest (ROIs) extracted from the CCTA series. The images are complemented by clinical features related to patients with/without CAD. For each patient, the dataset contains i) the original CCTA series, ii) the annotations related to EAT ROI, iii) the annotations related to PAT ROI detected around the anterior interventricular artery (IVA), and iv) some established clinical features highly predictive for CAD10. As a result, the dataset proposed here represents a multimodal dataset, namely MultiD4CAD (Multimodal Dataset for Coronary Artery Disease), that collects data relative to the CAD disease.
In addition to its significant clinical implications, the proposed dataset provides a valuable opportunity to address important data analysis challenges. CCTA images can be used for the extraction of radiomic features (e.g., enabling the training of shallow learning classifiers) or used for deep architectures training (e.g., Convolutional Neural Networks (CNNs), Visual Transformers (ViTs), etc.). Models based on this multimodal dataset - composed of clinical data and radiological images - have the potential to find hidden patterns between data from different sources, useful for improving the prediction of outcomes in CAD-affected patients. Moreover, the availability of clinical data opens methodological challenges related to multimodal data integration, enabling the implementation of high-performance Clinical Decision Support Systems (CDSSs).
The rest of the paper is organized as follows: Section ’Methods’ describes the procedures used in producing the data, Section ’Data Records’ explains data record proposed with this work, including the repository used to store the dataset, and provides an overview of the data files and their formats, Section ’Technical Validation’ presents the analyses performed to assess/support the technical quality of the dataset, Section ’Usage Notes’ contains the instructions necessary to assist researchers with the reuse of the data, finally Section ’Dataset and Code Availability’ indicates whether and under what conditions the code to obtain the data can be accessed.
Methods
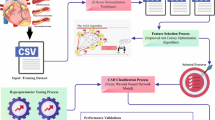
The proposed dataset was acquired following Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines11 to enhance standardization in the reporting procedure. The STROBE guidelines are recommendations designed to improve the quality and transparency of reporting in observational studies. These guidelines provide a checklist of items that should be included in reports of cohort, case-control, and cross-sectional studies to ensure clarity, accuracy, and completeness, aiding in the reproducibility and critical appraisal of research findings. The STROBE report concerning this study is available in Table S1 of the Supplementary Material. Figure 1 depicts the overall workflow showing the implemented steps to obtain the proposed MultiD4CAD.
Overall flow diagram showing the processing chain implemented to obtain MultiD4CAD, the multimodal dataset proposed with this study. Acronym specification: CAD: Coronary Artery Disease; CCTA: Coronary Computed Tomography Angiography; CDSS: Clinical Decision Support Systems; CT: Computed Tomography; EAT: Epicardial Adipose Tissue; PAT: Pericoronary adipose tissue; ROI: Region Of Interest. File format specification: csv: comma-separated values; dcm: DICOM; nii: NIfTI; txt: text.
Study Approval and Consent to Publish
A detailed report outlining the study’s objectives was submitted to the Ethics Committee of the University Hospital Policlinico ‘P. Giaccone’ of Palermo. Following a thorough evaluation, the Committee approved the proposed project (minutes N. 8/2024 of 03/19/2024 and N. 14/2025 of 06/09/2025). Written informed consent was waived because of the retrospective nature of the study. Conflict of interest was excluded, researchers furthermore declared to adhere to the rules of good clinical practice and to the Declaration of Helsinki. Furthermore, this agreement states that there is no restriction on the dissemination and publication of the study results, in compliance with the regulations in force concerning the confidentiality of sensitive data.
Data Inclusion and Exclusion criteria
Our dataset includes patients with suspected CAD who underwent CCTA exam at the University Hospital Policlinico ’P. Giaccone’. Initially, 300 patients were enrolled, but only 118 were selected considering the following exclusion criteria:
-
patients who have undergone coronary bypass surgery;
-
patients previously subjected to coronary stent implantation;
-
particularly tortuous anatomy of the anterior interventricular artery (IVA);
-
patients with cardiac pacemakers;
-
poor quality images (i.e., low opacification of coronary arteries, motion artifacts).
The presence of cardiovascular risk factors was assessed for each patient according to the Framingham Risk Score10. In particular, the following cardiovascular risk factors were considered: obesity (BMI>30), hypertension (blood pressure > 140/80 or need for antihypertensive therapy), diabetes mellitus, hypercholesterolemia (LDL cholesterol > 130 mg/dl or statin intake), family history of cardiovascular disease, and smoking habit. Figure 2 shows the diagram related to the exclusion criteria used during enrollment, aimed at the construction of the dataset.
CCTA Image Data Collection
The CCTA series were acquired through a 128-slice CT scanner SOMATOM Definition AS+ (Siemens Healthcare, Forchheim, Germany) provided by the Radiology Section of the University Hospital Policlinico ’P. Giaccone’. After the acquisition, collected images were transferred to a workstation equipped with dedicated diagnostic software for processing, specifically Syngo Via Cardiac (Siemens Healthcare, Forchheim, Germany), exploited to calculate the calcium score and to do MultiPlanar Reconstruction (MPR) to detect and evaluate plaques in coronary arteries. Image analysis was performed from October 2019 to January 2020.
The exam consists of an initial scan without the administration of a contrast agent, using a sequential technique to evaluate the calcium score. Subsequently, a post-contrast scan focused on the heart is performed using a restricted field of view (FOV). To supply the iodinated contrast (50-80 ml), an antecubital intravenous access site with at least a 20-gauge cannula was established. ECG-gated synchronized imaging was conducted to achieve a complete cardiac cycle acquisition. Image reconstruction occurred in 5-10% increments of the RR-interval, equivalent to 10-20 separate phases, with a slice thickness set at 0.7 mm for improved diagnostic accuracy with an interval of 0.4 mm.
Dataset De-identification
Any patient-related information/metadata contained in the DICOM header of the CCTA series was removed before starting the subsequent processing and analysis. This anonymization was performed using a custom-made tool, implemented using the Matlab IDE, compliant with DICOM supplement 142 (Clinical Trial De-identification Profiles)12. This step was mandatory to respect the patient’s privacy. In particular, data related to names, dates, times, comments, and center-related information were deleted. Instead, some DICOM attributes related to the patient (e.g., gender, age) and to the images (e.g., scanner type, pixel spacing, slice thickness) were maintained to preserve the statistical distribution of the dataset and the image metadata.
Clinical characteristics of the published data
The onset of chronic CAD can be influenced by various factors, and the relationship among them has been examined in numerous scientific studies. However, it is crucial to emphasize that correlation does not necessarily imply a direct causal relationship and multiple factors can contribute to the complex pathogenesis of chronic coronary artery disease. The Framingham Heart Study is one of the most famous long-term epidemiological studies on cardiovascular disease, through which it has been possible to identify risk factors for heart disease and to understand the patterns of onset and progression of cardiovascular disease. Over the years, the Framingham Heart Study has generated a vast amount of data that has significantly contributed to the understanding of the causes of heart disease. It has provided crucial insights into some risk factors such as hypertension, elevated cholesterol levels, smoking, diabetes, familiarity with cardiovascular diseases, and obesity10, included in this study.
To create a comprehensive multimodal dataset capable of supporting cardiovascular disease diagnosis, in addition to data from CCTA imaging, the following risk factors were also considered: arterial hypertension, hypercholesterolemia, smoke, diabetes, familiarity, and obesity. For completeness, the characteristics of sex and age features have been included, although they are sometimes considered confounding factors. The decision of whether to include these risk factors in defining the predictive model is left to the users of the dataset.
Clinical data were collected from the electronic patient record. A brief description of each clinical characteristic is provided below:
-
hypertension causes structural damage and endothelial dysfunction in the coronary arteries. PAT can amplify these effects through i) the secretion of pro-inflammatory cytokines that promote arterial inflammation, and ii) the impairment of endothelial cell function, facilitating the penetration of lipoproteins into the arterial walls13;
-
hypercholesterolemia, characterized by elevated levels of LDL cholesterol in the blood, is a known risk factor for CAD. PAT promotes the accumulation of lipids in the coronary arteries, facilitating the deposition of LDL cholesterol and altering the lipid profile by reducing HDL cholesterol levels and promoting the formation of atherosclerotic plaques14;
-
smoking is a powerful risk factor for CAD and influences PAT through direct damage to endothelial cells, impairing vascular function, and promoting systemic and local inflammation. Additionally, smoking is associated with increased LDL cholesterol and reduced HDL cholesterol, aggravating lipid accumulation in the coronary arteries;
-
diabetes (especially type 2) is associated with high blood glucose levels and insulin resistance. PAT interacts with diabetes by promoting systemic inflammation and insulin resistance, exacerbating the formation of atherosclerotic plaques. Chronic hyperglycemia damages endothelial cells, facilitating the adhesion of lipoproteins and plaque formation, a process amplified by PAT through the secretion of inflammatory cytokine;
-
a family history of cardiovascular diseases implies a genetic predisposition to risk factors such as hypertension, hypercholesterolemia, and inflammation. PAT can modulate the expression of genes involved in CAD, influencing the severity and age of onset of the disease. Genetic variants can influence the predisposition to inflammation and the formation of atherosclerotic plaques, a process in which PAT plays a significant role through its metabolic and inflammatory activity;
-
obesity plays a crucial role in the development of CAD through a combination of systemic inflammation, insulin resistance, lipid alterations, and elevated blood pressure, all factors that, as we have seen, promote the formation and progression of atherosclerotic lesions, characteristic of chronic coronary artery disease.
The final dataset includes 83 males and 35 females, with an overall average age of 60.41 ± 13.23. Regarding the class distribution, our dataset is composed of 78 samples for the ’with CAD’ class, and 40 samples for the ’without CAD’ class. Table 1 summarizes some statistics related to the clinical characteristics of the dataset.
Epicardial and Pericoronary Adipose Tissues Segmentation
ROI segmentations were performed by radiologists belonging to the research group with experience in annotation and quantification of cardiac adipose tissue. In particular, the segmentation and quantification of the two ROIs — containing the EAT and the PAT respectively — were performed through a custom-made automated software. The tool implements a semi-supervised procedure, in which the radiologist’s supervision relates to defining the range of CCTA slices to be processed and to setting the initial and final ROIs containing the adipose tissue. In particular, the main steps constituting the processing pipeline for EAT segmentation are15: i) ROI Selection; ii) ROI Interpolation; iii) ROI Refinement; iv) EAT Detection; v) Fat Volume and Quartiles Computation. Instead, the main steps constituting the processing pipeline for PAT segmentation are4: i) VOI containing the Anterior Interventricular Artery (IVA) selection; ii) reference ROIs (around the IVA) insertion; iii) automatic ROIs interpolation; iv) automatic PAT detection within slices in the range of interest. Using the mentioned software, ROI segmentations were performed by radiologists belonging to the research group with experience in annotation and quantification of cardiac adipose tissue. This software was used and validated in several other papers, aimed at studying i) the correlation between PAT and CAD4, ii) to evaluate the feasibility of texture analysis of EAT and thoracic subcutaneous fat (TSF) in patients undergoing CCTA14, and iii) variability of EAT quartiles distribution on patients under statin administration16.
To detect EAT and PAT regions properly, images belonging to the phase with the best quality and fewer motion artifacts (at the level of the proximal segment of the left anterior descending coronary artery) were selected. The segmentation software supports the radiologists through a user-friendly Graphical User Interface (GUI), in which a CCTA series is loaded and performs semi-automatically the detection of adipose tissues.
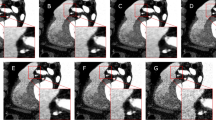
For all the implementation details about the tool, please visit the previous paper for EAT15 and PAT4 segmentation. Figures 3 and 4 show two segmentation examples of EAT and PAT, respectively. The figures have been scaled by a factor of 0.75 to improve the graphical visualization.
After delineation, the software converts and stores the CCTA images and the segmentation masks in NifTI (.nii) format. Raw DICOM data are converted to NIfTI format using the dcm2niix Python script (available at https://github.com/rordenlab/dcm2niix). The NifTI format was employed for optimized use both for CNNs training and for radiomic studies. Radiomic features can be extracted via appropriate tools (i.e., PyRadiomics17, an IBSI-compliant tool for radiomic features extraction freely available online18).
Data Records
The dataset is available at Zenodo19 (https://doi.org/10.5281/zenodo.15063615). Figure 5 visualizes the structure of directories and data contained in the proposed multimodal dataset. The structure is particularly intuitive: each of the 2 types of adipose tissue is contained in a specific folder (’Epicardial Adipose Tissue’ and ’Pericoronary Adipose Tissue’ respectively), within which all samples (i.e., the patients) are contained. Within a specific adipose tissue, each folder (e.g., ’P001’ and ’P118’ in Fig. 5) represents a specific sample, containing 2 NIfTI (.nii extension) files representing the images of the CCTA series and the corresponding segmentation masks. Moreover, there are 3 .csv files with i) the clinical data and the labels, ii) the metadata of the CT series, and iii) the labels alone.
Technical Validation
Clinical evaluation of the patients, assessment of CCTA series, and validation of clinical features were performed by radiologists with experience in CCTA reporting; in inconsistent assessment scenarios, a consensus was reached through discussion between the radiologists. Segmentation of epicardial and pericoronary adipose tissues was performed by radiologists with experience in annotation and quantification of cardiac adipose tissue. In particular, patients were examined during normal clinical routine. The examination was requested with a specific clinical indication (suspected chronic CAD), and the patients were analyzed from a clinical/radiological perspective by radiologists with experience in cardiac imaging. To ensure the quality and consistency of the dataset, rigorous exclusion criteria were established (see Fig. 2 in Data Inclusion and Exclusion criteria Section for further details). These measures were designed to maintain homogeneity among clinical cases and enhance the reliability of the collected data. Specifically, patients with prior clinical history (e.g., surgery intervention, stent implantation, anomalous IVA morphology, etc.) were excluded.
Application Scenarios
MultiD4CAD offers the potential to address a range of AI-related challenges. In particular:
Segmentation Assessment
EAT and PAT segmentation is a time-consuming and operator-dependent process. For this reason, it is desirable to have computer-assisted tools able to support this task. Semi-automatic segmentation of EAT and PAT has already been explored in15 and4, respectively. The availability of the validated annotations in our dataset enables the exploration of innovative methods, like deep architectures, for EAT and PAT segmentation in the CCTA series.
Classification Tasks
It is possible to use EAT and PAT annotations to support the diagnosis of CAD via supervised learning algorithms. In particular, multimodal models trained with imaging features extracted from PAT and clinical features were employed for CAD prediction4. In14 was evaluated the feasibility of texture analysis of EAT and the thoracic subcutaneous fat in patients undergoing CCTA. However, the mentioned works have not yet been integrated with features derived from EAT. For this reason, it opens several challenges for future developments.
Radiomic Studies
Through the validated EAT and PAT annotations, it is possible to implement radiomic analysis workflows optimized for CCTA. In4, an explainable radiomic workflow was implemented to find biomarkers correlated with the presence of CAD and a strong correlation between radiomic features and radiological visual patterns was found.
Deep Models Development
Considering the higher capability of deep architectures to extract informative features compared to shallow methods20,21, it is possible to investigate the use of deep architecture for segmentation and classification. This last aspect opens methodological challenges for training in scenarios with limited data availability.
Multimodal Learning
The dataset facilitates the integration of three distinct sources of information (PAT, EAT, and clinical features). This enables the exploration of methods that leverage intrinsic correlations among these sources to develop high-performing predictive models.
Usage Notes
The entire dataset, consisting of the images in .nii format regarding the EAT and PAT and their segmentation masks, and the .csv file with i) the clinical features and labels of the samples, and ii) the CT imaging metadata, are hosted on Zenodo, a controlled-access sharing platform.
The MultiD4CAD dataset (at the link https://doi.org/10.5281/zenodo.15063615) has been uploaded to Zenodo with restricted access19: only authenticated and authorized users can download it. Anyone wishing to access the dataset must follow the procedure:
-
Create an account on Zenodo, ensuring that either the username or the email address (or both) are public. If you already have a Zenodo account, make sure that at least one of these identifiers is public.
-
Complete and sign the Data Use Agreement (DUA)22 and send a signed copy at mailto:medicalimaginglab@unipa.it. You should put ‘MultiD4CAD Request access’ as the subject line, and in the email body specify the identifying information you made public when you activated your Zenodo account, i.e., username or email address.
-
On the Zenodo page of the dataset, you have to request access, specifying the first name, last name, affiliation, and email of the reference person who completed and sent the DUA (step 2).
-
Within two working days, you will be granted the ‘Reader’ access to the dataset -"MultiD4CAD: Multimodal Dataset composed of CT and Clinical Features for Coronary Artery Disease Analysis”. You can check the requests directly on Zenodo (‘My dashboard > Requests’). You should also receive a notification via email. Please check the ‘Other’ or ‘Spam’ folders if you do not see it.
-
Accept the request to join the Zenodo community, and you will have access to the dataset and download the data.
Acronyms
List of acronyms used in the study.
-
BMI: Body Mass Index
-
CAD: Coronary Artery Disease
-
CNNs: Convolutional Neural Networks
-
CCTA: Coronary Computed Tomography Angiography
-
CDSS: Clinical Decision Support System
-
CT: Computed Tomography
-
DICOM: Digital Imaging and COmmunications in Medicine
-
EAT: Epicardial Adipose Tissue
-
FOV: Field Of View
-
GUI: Graphical User Interface
-
HDL: high-density lipoproteins
-
HU: Hounsfield Units
-
IVA: Anterior Interventricular Artery
-
LDL: low-density lipoproteins
-
NIfTI: Neuroimaging Informatics Technology Initiative
-
PAT: Pericoronary Adipose Tissue
-
ROI: Region Of Interest
-
STROBE: Strengthening the Reporting of Observational Studies in Epidemiology
-
ViT: Visual Transformer
-
VOI: Volume Of Interest
Code availability
The custom-made software used for the detection of the epicardial and pericoronary adipose tissues is already freely available online at the link https://sites.unipa.it/medimglab/Software.html.
Further details on the implemented segmentation approaches of epicardial and pericoronary fat tissues can be found in the following two papers, respectively:
• Militello, C., Rundo, L., Toia, P., Conti, V., Russo, G., Filorizzo, C., Maffei, E., Cademartiri, F., La Grutta, L., Midiri, M., Vitabile, S. (2019). A semi-automatic approach for epicardial adipose tissue segmentation and quantification on cardiac CT scans. Computers in Biology and Medicine, 114, 103424. https://doi.org/10.1016/j.compbiomed.2019.103424
• Militello, C., Prinzi, F., Sollami, G., Rundo, L., La Grutta, L., Vitabile, S. (2023). CT radiomic features and clinical biomarkers for predicting coronary artery disease. Cognitive Computation, 15(1), 238-253. https://doi.org/10.1007/s12559-023-10118-7
References
Stone, G. W. et al. A prospective natural-history study of coronary atherosclerosis. New England Journal of Medicine 364, 226–235 (2011).
Sacks, H. S. & Fain, J. N. Human epicardial adipose tissue: a review. American heart journal 153, 907–917 (2007).
Mahabadi, A. A. et al. Association of epicardial fat with cardiovascular risk factors and incident myocardial infarction in the general population: the heinz nixdorf recall study. Journal of the American College of Cardiology 61, 1388–1395 (2013).
Militello, C. et al. CT radiomic features and clinical biomarkers for predicting coronary artery disease. Cognitive Computation 15, 238–253 (2023).
Li, M. et al. Association of pericardiac adipose tissue with coronary artery disease. Frontiers in Endocrinology 12, 724859 (2021).
Marwan, M. & Achenbach, S. Quantification of epicardial fat by computed tomography: why, when and how? Journal of cardiovascular computed tomography 7, 3–10 (2013).
Oikonomou, E. K. et al. A novel machine learning-derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography. European heart journal 40, 3529–3543 (2019).
Obaid, D. R. et al. Atherosclerotic plaque composition and classification identified by coronary computed tomography: assessment of computed tomography–generated plaque maps compared with virtual histology intravascular ultrasound and histology. Circulation: Cardiovascular Imaging 6, 655–664 (2013).
La Grutta, L. et al. Quantification of epicardial adipose tissue in coronary calcium score and CT coronary angiography image data sets: comparison of attenuation values, thickness and volumes. The British Journal of Radiology 89, 20150773 (2016).
D’Agostino Sr, R. B. et al. General cardiovascular risk profile for use in primary care: the framingham heart study. Circulation 117, 743–753 (2008).
Von Elm, E. et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. International journal of surgery 12, 1495–1499 (2014).
DICOM Standards Committee. Supplement 142-"clinical trial de-identification profiles. DICOM Standard 1–44 (2011).
Weber, T. et al. Hypertension and coronary artery disease: epidemiology, physiology, effects of treatment, and recommendations: A joint scientific statement from the austrian society of cardiology and the austrian society of hypertension. Wiener Klinische Wochenschrift 128, 467–479 (2016).
Agnese, M. et al. Epicardial and thoracic subcutaneous fat texture analysis in patients undergoing cardiac CT. Heliyon 9 (2023).
Militello, C. et al. A semi-automatic approach for epicardial adipose tissue segmentation and quantification on cardiac CT scans. Computers in Biology and Medicine 114, 103424 (2019).
Toia, P. et al. Epicardial adipose tissue changes during statin administration in relation to the body mass index: A longitudinal cardiac CT study. Applied Sciences 13, 10709 (2023).
van Griethuysen, J. et al. Computational radiomics system to decode the radiographic phenotype. Cancer research 77, e104-e107 (2017).
PyRadiomics website. https://pyradiomics.readthedocs.io/en/latest/. Accessed on October 16, 2024.
Prinzi, F. et al. MultiD4CAD: Multimodal dataset composed of ct and clinical features for coronary artery disease analysis. https://doi.org/10.5281/zenodo.15063615 (2025).
Truhn, D. et al. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology 290, 290–297 (2019).
Sun, Q. et al. Deep learning vs. radiomics for predicting axillary lymph node metastasis of breast cancer using ultrasound images: don’t forget the peritumoral region. Frontiers in oncology 10, 53 (2020).
Prinzi, F. et al. DUA - MultiD4CAD: Multimodal dataset composed of ct and clinical features for coronary artery disease analysis https://doi.org/10.5281/zenodo.15063937 (2025).
Acknowledgements
This research was funded by: i) the Italian Complementary National Plan PNC-I.1 “Research initiatives for innovative technologies and pathways in the health and welfare sector” D.D. 931 of 06/06/2022, “DARE - DigitAl lifelong pRevEntion” initiative, code PNC0000002, CUP: B53C22006460001; ii) the European Union - Next Generation EU - Progetti di Ricerca di Rilevante Interesse Nazionale (PRIN) 2022, Prot. 2022ENK9LS. Project: “EXEGETE: Explainable Generative Deep Learning Methods for Medical Image and Signal Processing”, CUP: B53D23013040006.
Author information
Authors and Affiliations
Contributions
F.P. and C.M. prepared the files composing the dataset; C.M. implemented the semi-automatic segmentation software able to detect the EAT and the PAT; L.L.G. and P.T. evaluated the patients clinically; G.S. and P.T. performed EAT and PAT image analysis; L.L.G supervised and verified the delineation results; F.P. and C.M. dealt with the anonymization and organization of the dataset; F.P. and C.M. wrote the manuscript; L.L.G. managed the procedure with the Local Ethics Committee for the study approval; L.L.G. and S.V. coordinated and supervised the entire study. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Prinzi, F., Militello, C., Sollami, G. et al. MultiD4CAD: Multimodal Dataset composed of CT and Clinical Features for Coronary Artery Disease Analysis. Sci Data 12, 1570 (2025). https://doi.org/10.1038/s41597-025-05743-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05743-w