Abstract
Accurate segmentation of pulmonary structures from computed tomography (CT) is critical for lung disease management, yet progress is hampered by a lack of large-scale, public datasets with comprehensive multi-structure annotations. To address this, we present the Airway and Pulmonary Vessel Structural Representation in CT (AirRC) dataset, comprising 254 CT scans from the LUNA16 dataset meticulously annotated with 3D masks for pulmonary veins, arteries, airway lumen, and airway wall. Technical validation was performed via 5-fold cross-validation using a custom MONAI-based deep learning pipeline. The model achieved high mean Dice Similarity Coefficients (DSC) for Pulmonary Veins (0.953), Pulmonary Arteries (0.950), and Airway Lumen (0.941), with strong performance on the challenging Airway Wall (0.866). A two-stage refinement strategy further improved small airway branch segmentation. External validation on public benchmarks (ATM’22, Parse2022, HiPas) confirmed the utility and generalizability of models trained on AirRC, establishing it as a robust resource for developing and evaluating advanced pulmonary segmentation algorithms.
Similar content being viewed by others
Background & Summary
Computed tomography (CT) plays a vital role in pulmonary imaging, serving as an indispensable tool in the diagnosis and study of lung diseases. Its ability to capture comprehensive chest images at nearly isotropic, sub-millimeter resolution during a single breath-hold makes CT exceptionally suited for visualizing the complex biological structures of the lungs, particularly the pulmonary vascular tree. Advances in CT technology have dramatically enhanced the ability to delineate fine pulmonary structures and lesions, making it an essential modality in pulmonary research and clinical practice.
In medical research and clinical management, particularly for respiratory diseases, accurate segmentation of pulmonary structures is critical. For instance, precise delineation of pulmonary arteries and veins provides key insights into the complex anatomical and pathological relationships between lung lesions and the vascular system, a capability particularly valuable in oncology and surgical planning1,2,3,4. It also enhances the evaluation of conditions such as pulmonary embolism and pulmonary hypertension5. Similarly, the precise segmentation of the airway lumen and wall provides critical, objective data for managing chronic obstructive pulmonary disease (COPD) and other airway remodeling diseases6,7,8.
Manual segmentation of these intricate structures is time-consuming, labor-intensive, and prone to variability, making it impractical for large-scale analysis1,9,10. Consequently, deep learning models have emerged as the most effective tools for automated segmentation10,11,12,13. However, the performance of these advanced models is fundamentally dependent on the availability of large-scale, accurately annotated public datasets, which remains a critical barrier in the field.
The academic community has recently made significant strides in addressing this data scarcity. For instance, the groundbreaking HiPaS study by Chu et al. demonstrated high-fidelity artery-vein segmentation on non-contrast CT—a task previously considered infeasible—by training on a large-scale, multi-center private dataset and highlighted the potential for non-invasive vascular analysis10. Similarly, Cheng et al. developed a novel fusion-based segmentation approach for lung cancer surgery planning and released an associated expert-annotated dataset titled “Pulmonary Arteries and Veins Segmentation in Lung”14. These contributions are invaluable and have significantly advanced the field’s capabilities.
However, a specific and critical need persists for a public dataset that is (1) built upon a widely-used, pre-existing benchmark, facilitating direct comparison and integration into existing research pipelines, and (2) provides comprehensive annotations for both the airway and vascular trees simultaneously, enabling the development of holistic, multi-task models. While datasets like HiPaS are powerful, their use requires researchers to adopt a new, standalone data collection. Furthermore, most existing datasets focus narrowly on either vessels or airways.
To fill this specific gap, we have developed and are releasing a novel dataset, termed Airway and Pulmonary Vessel Structural Representation in CT (AirRC). The unique contribution of AirRC lies in providing comprehensive annotations for a substantial subset (254 scans) of the LUNA16 (Lung Nodule Analysis 2016) challenge dataset—a cornerstone benchmark in the pulmonary imaging community. Our detailed, manually refined annotations provide three-dimensional segmentation masks for four key structures: pulmonary veins, pulmonary arteries (both extended into the mediastinum for anatomical completeness), airway lumen, and airway wall. By anchoring these rich, multi-structure annotations to a familiar and universally accessible baseline dataset, AirRC provides a robust and ready-to-use resource for the research community. We aim to foster the development, evaluation, and benchmarking of advanced AI models for precise and comprehensive pulmonary structure segmentation, thereby driving innovation in both pulmonary imaging research and its clinical applications.
Comparing with other datasets
To contextualize the specific contributions of AirRC, it is instructive to compare its features with other prominent publicly available datasets for pulmonary segmentation, a detailed summary of which is provided in Supplementary Table 1.
The field’s foundational datasets, while pioneering, present significant limitations for training modern, data-hungry deep learning models. The VESSEL12 challenge, for instance, provides only sparse, point-based annotations where labels exist only for voxels agreed upon by three annotators, rather than dense, complete segmentation masks suitable for training15. Similarly, the CARVE14 dataset, while focused on artery-vein separation, is limited by its annotation strategy; full vascular trees were annotated in only 10 of the 55 non-contrast CT scans, leaving the majority of the dataset incompletely labeled1.
More recent high-quality datasets are often highly specialized, providing excellent annotations for a single anatomical structure. The ATM’22 (Airway Tree Modeling 2022) challenge released a large-scale (500 scans) dataset with meticulously refined airway annotations, making it a benchmark for airway-specific tasks. However, it does not include any vascular structures16. Conversely, the Parse2022 challenge provided a valuable resource of 100 contrast-enhanced CTPA scans with multi-expert consensus annotations, but these are exclusively for the pulmonary arteries, omitting the venous system entirely17.
Other datasets present unique usability challenges for standard image-to-mask segmentation tasks. The TotalSegmentator dataset’s vessel annotations were generated by a classical, non-deep-learning automated algorithm rather than by human experts, and it combines arteries and veins into a single class, precluding studies that require physiological differentiation18. The innovative Pulmonary-Tree-Labeling (PTL) dataset contains detailed, 19-class branch-level labels for airways, arteries, and veins, but crucially, it does not provide the corresponding raw CT image data, making it suitable for graph-based analysis but not for training traditional image segmentation models19.
Even the most recent and advanced datasets, while powerful, serve distinct purposes. The groundbreaking HiPaS dataset demonstrated high-fidelity artery-vein segmentation on both contrast and non-contrast CT, backed by meticulous manual annotations on a large private cohort. However, it is a standalone resource focused exclusively on the vasculature, and only a subset of its annotated data (n = 250) has been publicly released10. The “Pulmonary Arteries and Veins Segmentation in Lung” dataset provides expert surgeon annotations for lung cancer surgery planning. However, its utility as a general-purpose benchmark is limited, as the public release consists of only 73 hemithorax cases, the annotations are often partial (covering only one lung), and the provided CT data consists of cropped patches with transformed intensity values, not full-field-of-view scans in original Hounsfield Units14.
AirRC distinguishes itself by occupying a unique and complementary niche within this landscape. It is the first public resource to provide comprehensive, co-registered, and expert-validated annotations for the complete airway and pulmonary vascular trees (arteries, veins, airway lumen, and airway wall) simultaneously. Critically, these rich, multi-structure annotations are layered directly onto 254 scans from the LUNA16 benchmark, a dataset already familiar to and widely used by the pulmonary research community. This design makes AirRC an ideal and ready-to-use resource for developing and validating holistic, multi-task segmentation models on a universally accessible baseline.
Methods
Rationale and selection of CT scans for annotation
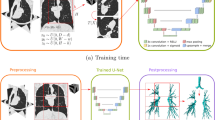
This study aimed to develop and publicly share a meticulously annotated dataset of pulmonary airways (distinguishing lumen and wall), and pulmonary vasculature (differentiating arteries and veins). To this end, candidate CT scans were drawn from the publicly available LUNA16 dataset (Fig. 1a). The original LUNA16 CT images are available under a Creative Commons Attribution 4.0 International (CC BY 4.0) license20,21 and can be obtained from their official distribution channels.
Study Workflow and Annotation Example. (a) Diagram illustrating the study workflow. (b) Axial view of the final annotation results. (c) Three-dimensional reconstruction of the annotated structures. In both (b) and (c), pulmonary veins are marked in blue, pulmonary arteries in red, airway lumen in green, and airway wall in yellow. (CV: Computer Vision).
The selection and annotation process, detailed below, was designed to maximize dataset quality and mitigate known biases from the parent LUNA16 collection. This process involved a multi-stage, human-in-the-loop workflow that ultimately yielded a final cohort of 254 high-quality annotated scans. One of the goals during the final case selection was to improve the balance of scanner manufacturers to enhance data diversity. The final manufacturer distribution of the resulting dataset is 142 cases from GE Medical Systems, 73 from Siemens, and 39 from Philips.
Image preprocessing
All selected CT images from the LUNA16, originally in MHD (MetaImage Header) format, were converted to the NIfTI (Neuroimaging Informatics Technology Initiative) format. The images were subsequently resampled to an isotropic spacing of 1 mm × 1 mm × 1 mm to ensure uniformity for all subsequent processing and annotation.
Annotation protocol for pulmonary airways and vasculature
A comprehensive, multi-stage annotation protocol was implemented for all selected scans to delineate pulmonary vasculature (arteries and veins) and the airway tree (lumen and wall). This protocol was designed to ensure both high efficiency and exceptional quality by leveraging a human-in-the-loop approach, which was crucial for maximizing annotation consistency across the entire dataset. The workflow consisted of four main stages:
Stage 1: Initial coarse segmentation using an automated pipeline
To expedite the manual annotation process, initial coarse segmentation labels for pulmonary vasculature and airways were automatically generated from the raw CT scans. This was performed using an in-house script developed in Python (v3.11), leveraging core libraries including SimpleITK (v2.3.1), OpenCV (v4.10.0), and Scikit-image (v0.24.0).
-
Pulmonary Vessel Coarse Segmentation: A 2D slice-wise approach was employed. Each axial slice underwent preprocessing, including clamping Hounsfield Unit values to the range of [−2000, 2000] and normalization to a 0–255 grayscale range, while preserving original spatial information. An initial lung field approximation was obtained via Otsu’s thresholding. This mask was then refined using a secondary threshold, calculated by multiplying the mean intensity of pixels above the Otsu threshold by a lung refinement factor (default: 0.75). Further refinements included clearing border pixels, applying adaptive median filtering, removing border-connected components, filling holes via a flood-fill algorithm, and smoothing using adaptively sized morphological erosion and dilation operations. To identify vessel candidates within these refined lung regions, linear contrast enhancement (factor: 1.5) was applied to the normalized slice. After temporarily masking out the lung regions, the mean intensity of the remaining non-lung pixels was computed. A final adaptive vessel threshold was derived by multiplying this mean intensity by a vessel adjustment factor (default: 1.25). Applying this threshold to the contrast-enhanced image yielded binary masks of potential vessel structures for each slice, which were subsequently combined into a 3D volume.
-
Airway Coarse Segmentation: A 3D seed-based region growing strategy was utilized. Initially, CT HU values were clamped to the range of [−1000, 0] to enhance airway visibility. A seed point was typically placed manually or semi-automatically within the superior portion of the trachea on this clamped volume. A 3D region growing algorithm, specifically SimpleITK’s ConfidenceConnectedImageFilter, was initiated from this seed. This algorithm automatically defined an intensity range based on local statistics around the seed point and grew iteratively into connected voxels falling within this dynamically determined range, governed by a sensitivity multiplier parameter (default used: 3.0). The resulting raw airway mask was then refined using 3D morphological opening and closing operations (kernel radius: 1 voxel) to remove small spurious regions and smooth the boundaries, yielding a single binary mask representing the connected airway tree without specific branch-level distinctions.
This automated pipeline was explicitly designed to produce initial, coarse segmentations suitable for subsequent expert review. While default values were generally applied for the lung refinement factor (0.75) and vessel adjustment factor (1.25) in vessel segmentation, these could be adjusted on a case-by-case basis if deemed necessary by the operator. Similarly, for airway segmentation, the crucial seed point placement was inherently case-specific, and parameters controlling region growing sensitivity and morphological post-processing were also adjusted when necessary to optimize the initial segmentation. This flexibility prioritized generating a usable initial segmentation over strict adherence to fixed parameters across all cases. It must be emphasized that this automated stage served solely as an efficiency tool to reduce manual labor; definitive anatomical accuracy for both vessels and airways was established exclusively through the subsequent expert manual annotation process.
Stage 2: First-pass manual refinement
Following the generation of these initial coarse labels, a rigorous manual review and refinement workflow was implemented to establish the final ground truth annotations. This critical step was performed by two human observers: Observer 1, with 5 years of clinical experience in thoracic radiology, and Observer 2, with 11 years of clinical experience in pulmonary imaging. Using 3D Slicer22 (v5.2.2+), the observers first overlaid the coarse segmentation masks onto the original CT images. They adjusted the CT display using standard lung window presets (e.g., window width 1400 HU, window level −500 HU) to optimally visualize the relevant structures.
Based on anatomical features, they meticulously classified vessels as either pulmonary arteries or veins and refined the boundaries of both vessels and the unified airways using interactive brush and eraser tools available in 3D Slicer.
This refinement process followed a two-stage verification protocol. Initially, Observer 1 performed the primary corrections and classifications. Subsequently, Observer 2 reviewed these modifications, providing further corrections and verification. Any disagreements between the two observers were resolved through consultation with a third senior observer possessing 15 years of experience in thoracic imaging, ensuring consistency and accuracy across all annotations.
Furthermore, recognizing the clinical significance of distinguishing the airway lumen from the airway wall, particularly for COPD and asthma, a specific sub-protocol was implemented to achieve this level of granular detail within the airway annotations. After the overall airway mask was finalized by the observers, it was loaded for further processing. A 3D binary erosion operation, implemented using the SimpleITK library, was applied to the mask using a spherical structuring element with a radius of 1 pixel. This morphological operation effectively shrank the segmented region by removing the outermost layer of voxels, thereby providing an approximation of the airway lumen. Subsequently, the airway wall was isolated by performing a pixel-wise subtraction of this derived lumen mask from the original, finalized airway mask. This derived lumen/wall separation underwent the same rigorous two-stage review and final arbitration process by the clinical experts to ensure the anatomical plausibility and accuracy of these finer details.
Stage 3: Intermediate model training and inference
The refined labels from Stage 2 were used to train a standard 3D deep learning segmentation model (based on the nnU-Net framework v2.5.1). This trained model was then used to perform inference on the entire pool of candidate CT scans from the LUNA16 dataset. The resulting predictions served as a new, high-quality, and anatomically consistent set of draft annotations.
Stage 4: Quality control, case selection, and final expert verification
This was the final and most critical stage for establishing the ground truth. First, all draft annotations from Stage 3 underwent a quality control review. Cases with significant, uncorrectable errors were excluded. From the remaining high-quality candidates, a final cohort was strategically selected. The decision to finalize the dataset at 254 scans was made to strike a deliberate balance: this scale is substantial enough to train robust deep learning models, yet manageable enough to allow for the extremely time-consuming, multi-expert verification required for the complex airway and vessel structures. The final selection was guided by the dual goals of (1) ensuring broad anatomical variability and (2) improving the balance of scanner manufacturers. The selected draft annotations were then provided to our two primary human observers for meticulous final correction and vessel classification, with any disagreements resolved by a third senior observer. The airway wall was subsequently derived from the finalized airway mask and underwent the same rigorous three-expert verification process.
The 254 cases that successfully passed this entire quality-controlled pipeline constitute the final AirRC dataset. This human-in-the-loop methodology was crucial for creating a standardized and high-quality ground truth. Figure 1b,c show an example of the final annotation.
Data Records
The primary data record consists of detailed, multi-structure annotations for a curated subset of 254 CT scans from the LUNA16 dataset. These annotations are publicly accessible via Figshare (https://doi.org/10.6084/m9.figshare.26878867)23 under a CC-BY 4.0 license. The Figshare repository contains the annotation files (.nii.gz) and a detailed metadata spreadsheet (metadata.xlsx). The original LUNA16 CT images must be obtained separately from their official distribution channels20,21. These original LUNA16 data sources are governed by a CC BY 4.0 license. Our provided annotations are designed to be used in conjunction with these original CT scans. A complete description of the metadata fields is provided in Supplementary Table 2. Each.nii.gz annotation file is a 3D volume spatially aligned with its corresponding CT scan (resampled to 1 mm isotropic spacing), with integer voxel values representing: 1 for Airway Lumen, 2 for Airway Wall, 3 for Pulmonary Arteries, and 4 for Pulmonary Veins. All other voxels have a value of 0.
Technical Validation
The quality and utility of the AirRC dataset were demonstrated through a comprehensive technical validation. A state-of-the-art segmentation framework was trained and evaluated using a rigorous 5-fold cross-validation scheme directly on the 254 cases of the AirRC dataset. The model’s generalizability was subsequently assessed on three independent, public benchmarks: ATM’22, Parse2022, and HiPas.
Segmentation framework and implementation details
Our segmentation pipeline was developed using MONAI (v1.5.0) and PyTorch Lightning (v2.5.2), adapting the core, state-of-the-art strategies popularized by the nnU-Net framework24, such as dynamic data preprocessing, comprehensive data augmentation, and sliding-window inference with Gaussian weighting. The network architecture was a 3D U-Net featuring residual blocks and deep supervision from four decoder levels. Technical validation was performed using a 5-fold cross-validation with stratification by scanner manufacturer to ensure robust and unbiased results. All models were trained using an SGD optimizer with a polynomial learning rate scheduler.
Two-stage refinement for airway lumen segmentation
To specifically address the well-known challenge of segmenting fine, distal airway branches, we adopted and implemented a two-stage refinement strategy. This approach is based on the winning solution from Team timi in the ATM’22 Challenge16, effectively leveraging hard case mining and a specialized loss function to enhance performance. Our goal was not to introduce a novel segmentation method, but rather to use this validated, state-of-the-art approach to rigorously assess the quality and utility of our AirRC dataset for this demanding task.
Stage 1 (Baseline Model)
A baseline model was trained for 1000 epochs on full CT volumes. The training objective was a Deep Supervision Dice and Cross-Entropy Loss. This composite loss, \({{\mathscr{L}}}_{\text{DiceCE}}\), combines a soft Dice loss (\({{\mathscr{L}}}_{\text{Dice}}\)) and a standard Cross-Entropy loss (\({{\mathscr{L}}}_{\text{CE}}\)), and is applied across all network outputs to establish a strong initial segmentation. The total loss for a single output is defined as:
For our network with S = 5 outputs (one final, four from deep supervision levels), the total training objective, \({{\mathscr{L}}}_{\text{Stage}1}\), is a weighted sum over all outputs:
where Ps is the prediction from the s-th output, G is the ground truth label, and ws are the supervision weights, which are set to decay exponentially (\({w}_{s}=1/{2}^{s}\)) and are normalized to sum to one, with the weight for the highest-resolution supervision layer set to zero. This strategy ensures robust gradient flow throughout the network’s depth.
Stage 2 (Refinement Model)
To specifically address the challenge of segmenting fine, distal airway branches, we adopted the two-stage refinement methodology proposed by Team timi in their winning solution to the ATM’22 challenge16. This involved training a second model for a more focused 150 epochs. Following their work, the training data for this stage consisted of image patches extracted from “hard case” regions (e.g., areas of prediction error) identified by the baseline model.
The core of this stage is a custom, class-weighted loss function, also following the design of Team timi16. The loss for a single output, \({{\mathscr{L}}}_{\text{Stage2\_base}}\), is a weighted sum over all foreground classes:
where \({\lambda }_{c}\) is the weight for class c, and \({{\mathscr{L}}}_{c}\) is the specific loss function for that class. The class weighting scheme set a weight of \({\lambda }_{\text{lumen}}=1.0\) for the airway lumen and \({\lambda }_{\text{other}}=0.5\) for all other classes to concentrate the model’s learning.
For the airway lumen, a specialized timi Loss (\({{\mathscr{L}}}_{\text{Timi}}\)) was used. It integrates a Focal Union component (\({{\mathscr{L}}}_{\text{FU}}\)), a technique and formulation introduced by Zheng et al.25, Zhang et al.26 and Team timi16 to better penalize false negatives at the periphery of the airway tree:
In our implementation, the weights were set to α = 0.5 and β = 1.0. This entire class-weighted loss structure was then applied across the deep supervision outputs using the same weighted summation as in Stage 1, effectively focusing the model’s capacity on improving airway segmentation fidelity.
Inference
During inference, a sliding-window approach with Gaussian importance weighting was employed using MONAI’s functionalities to generate smooth and consistent predictions across entire CT volumes. For final evaluation on external datasets, an ensemble of the five models from the cross-validation was used.
The model’s performance was assessed using a comprehensive set of metrics. For all segmented structures, we calculated standard voxel-based metrics: the Dice Similarity Coefficient (DSC) for volumetric overlap, Recall (Sensitivity) to measure the fraction of ground truth found, and Precision to measure the fidelity of the predictions. We also calculated the False Positive Rate (FPR) to assess model specificity. Consistent with expectations for high-volume 3D segmentation tasks with extreme class imbalance, the resulting FPR values were consistently negligible across all experiments (universally < 0.001). As this metric did not provide sufficient discriminatory power between models or cases, it is not presented in our results tables to maintain clarity.
Recognizing that voxel-based metrics may not fully capture the anatomical fidelity of complex branching structures, we included two specialized topological metrics for the airway lumen. This follows the methodology adopted by the ATM’22 challenge. These metrics are:
Branch Detected Rate (BDR): This measures the percentage of correctly detected branches from the ground-truth airway tree, where a predicted branch is considered ‘correct’ only if more than 80% of its length overlaps with a ground-truth branch.
where \({B}_{\det }\) is the number of correctly detected branches and \({B}_{\text{ref}}\) is the total number of branches in the ground-truth.
Detected Length Rate (DLR): This measures the fraction of the total length of the ground-truth airway tree that is detected by the segmentation. It is calculated as:
where \({T}_{\det }\) is the total length of all correctly detected branch centerlines and \({T}_{\text{ref}}\) is the total length of all branch centerlines in the ground-truth.
Together, this comprehensive suite of metrics provides a robust evaluation of both volumetric accuracy and topological completeness.
Internal validation on the AirRC dataset
The aggregated performance across all 254 cases in the 5-fold cross-validation is summarized in Table 1. The results confirm that AirRC’s annotations are of high quality and consistency, capable of training robust and accurate deep learning models.
The framework achieved excellent segmentation performance for major vascular and airway structures. The mean DSC was consistently high for Pulmonary Veins (0.953 ± 0.010), Pulmonary Arteries (0.950 ± 0.008), and the Airway Lumen (0.941 ± 0.022). Critically, the exceptionally low standard deviations observed for these major structures, even across a diverse 254-case cohort, serve as strong evidence of the high quality and consistency of the underlying annotations in the AirRC dataset. The anatomical fidelity of the Airway Lumen segmentation was further confirmed by outstanding topological metrics: a mean BDR of 0.978 ± 0.017 and a DLR of 0.989 ± 0.006, indicating that nearly the entire airway tree structure was correctly captured.
Segmentation of the Airway Wall yielded a strong mean DSC of 0.866 ± 0.046. As expected, this result is lower than that of the lumen, which accurately reflects the inherent challenge of delineating thin anatomical structures that are highly susceptible to partial volume effects in CT imaging.
External validation
To assess the generalizability of models trained on AirRC, the ensemble model was evaluated on the ATM’22, Parse2022, and HiPaS datasets (Table 2).
For airway segmentation on the ATM’22 dataset, a preliminary technical review of the 300 validation scans was conducted to ensure data integrity. This review identified 10 scans with a non-standard intensity format (positive integers instead of Hounsfield Units) and one scan with a significant label-to-image spatial misalignment. As these 11 cases presented objective technical incompatibilities, they were excluded from our analysis. On the remaining curated set of 289 scans, the model achieved a mean DSC of 0.821 ± 0.044 for the Airway Lumen. The remaining performance gap compared to our internal validation can be largely attributed to a mismatch in annotation granularity, a pattern observed across many cases. As visually demonstrated by a representative example in Fig. 2a,b, our model, trained on highly detailed AirRC annotations, consistently segments finer peripheral branches that are not labeled in the ATM'22 ground truth. In the evaluation, these anatomically plausible detections are penalized as False Positives, which lowers the overall Precision and DSC. While the model performed well on average, we also investigated the cause of the lower minimum DSC (0.522). This was found to be an extreme instance of the same pattern, where a particularly pronounced mismatch in annotation detail on an outlier case led to a significantly lower score. Despite this, the model’s high overall Recall (0.977) and excellent topological scores (BDR of 0.987, DLR of 0.988) confirm that it successfully identified the near-complete airway tree structure as defined by the ATM'22 annotations across the vast majority of cases.
Visual Analysis of Annotation Style Differences on External Datasets. This figure compares segmentation results from our AirRC-trained model against the reference annotations from the ATM'22 and HiPas datasets to illustrate differences in labeling philosophy. The color scheme highlights these differences: blue indicates regions segmented only by our model (reflecting our annotation style); red indicates regions labeled only in the external dataset’s ground truth; and yellow represents the consensus on the core anatomical structures. Panels (a,b) show an Airway Lumen from the ATM'22 dataset in coronal and 3D views. Panels (c,d) and (e,f) show the Pulmonary Arteries and Veins, respectively, from a HiPas case. In all instances, the predominance of yellow demonstrates a strong foundational agreement on the main structures. The key observation is the consistent pattern of blue voxels at the distal ends of the vascular and airway trees. This visually confirms that our model, trained on the detailed AirRC annotations, consistently identifies more peripheral, finer-caliber branches. While these are anatomically plausible, they are penalized as False Positives when evaluated against a reference standard with a different scope of annotation, which explains the performance gap observed in our external validation.
For pulmonary vessel segmentation, the model demonstrated robust generalization to external contrast-enhanced CTPA datasets. On Parse2022, it achieved a mean DSC of 0.802 ± 0.035 for Pulmonary Arteries. Similarly, after excluding one HiPas case with a critical image-to-label orientation mismatch, performance on the curated HiPas dataset was also strong, reaching a mean DSC of 0.816 ± 0.041 for Pulmonary Arteries and 0.786 ± 0.042 for Pulmonary Veins.
The ability to achieve DSC scores above 0.80 on these external benchmarks is a strong indicator of the model’s generalization capability. This is significant as the model was trained exclusively on the AirRC (LUNA16) cohort and applied in a zero-shot setting to these new clinical domains, which differ in patient populations, scanner protocols, and acquisition indications.
The remaining performance gap compared to our internal validation is attributable to a combination of this expected domain shift and subtle variations in annotation philosophy. This difference in annotation style is visually demonstrated for a representative HiPas case in Fig. 2(c–f). A review of such cases reveals that our model, reflecting the detailed AirRC annotation protocol, often segments finer distal vessels than are labeled in the external ground truth. These anatomically plausible detections are penalized as False Positives, explaining the performance difference and highlighting subtle but important variations in annotation criteria across datasets.
Overall, the strong performance in both internal and external validations underscores the utility and quality of the detailed, multi-structure annotation protocol employed in creating the AirRC dataset, confirming it as a valuable public resource for training and validating high-performing, generalizable segmentation models.
Limitations
While the AirRC dataset provides detailed multi-structure annotations, several limitations should be acknowledged. Firstly, while the dataset size of 254 scans is substantial, training highly complex deep learning models may still benefit from pre-training or transfer learning strategies. Secondly, the dataset inherits characteristics from its LUNA16 source, including a potential focus on nodule-positive cases and missingness in some clinical metadata. Thirdly, while our protocol minimized variability, manual segmentation is inherently subject to some level of expert interpretation. Fourthly, our technical validation represents one specific high-performance implementation; outcomes could differ with other architectures. Finally, the external validation underscored the challenges of domain shift across datasets with differing imaging protocols and annotation standards. Furthermore, our decision to resample all CT scans to a uniform 1 × 1 × 1 mm3 isotropic resolution standardizes model training but may limit the adaptability of the dataset to future CT technologies with higher native resolutions and could affect fine details in sub-millimeter structures, such as distal airways.
Usage Notes
Before using our annotated data, researchers are required to resample the original CT images to a voxel size of 1 mm × 1 mm × 1 mm to ensure dimensional compatibility.
Data availability
The AirRC dataset generated in this study has been deposited in the Figshare repository under accession code [https://doi.org/10.6084/m9.figshare.26878867].
Code availability
The primary training and inference code used in this study is publicly available on GitHub and can be accessed at https://github.com/b-niu/AirRC.
References
Charbonnier, J. P. et al. Automatic Pulmonary Artery-Vein Separation and Classification in Computed Tomography Using Tree Partitioning and Peripheral Vessel Matching. IEEE Trans Med Imaging 35, 882–892, https://doi.org/10.1109/TMI.2015.2500279 (2016).
Jimenez-Carretero, D. et al. A graph-cut approach for pulmonary artery-vein segmentation in noncontrast CT images. Med Image Anal 52, 144–159, https://doi.org/10.1016/j.media.2018.11.011 (2019).
Pu, J. et al. Automated identification of pulmonary arteries and veins depicted in non-contrast chest CT scans. Med Image Anal 77, 102367, https://doi.org/10.1016/j.media.2022.102367 (2022).
Wang, L. et al. Evaluating efficacy and safety of a novel registration-free CT-guided needle biopsy navigation system (RC 120): A multicenter, prospective clinical trial. Lung Cancer 198, 108025, https://doi.org/10.1016/j.lungcan.2024.108025 (2024).
Nardelli, P. et al. Pulmonary Artery-Vein Classification in CT Images Using Deep Learning. IEEE Trans Med Imaging 37, 2428–2440, https://doi.org/10.1109/TMI.2018.2833385 (2018).
Kurashima, K. et al. Changes in the airway lumen and surrounding parenchyma in chronic obstructive pulmonary disease. 523–532 (2013).
Ortiz-Puerta, D., Diaz, O., Retamal, J., Hurtado, D. E. J. F. I. B. & Biotechnology. Morphometric analysis of airways in pre-COPD and mild COPD lungs using continuous surface representations of the bronchial lumen. 11, 1271760 (2023).
Walters, E. H., Shukla, S. D., Mahmood, M. Q. & Ward, C. J. E. R. R. Fully integrating pathophysiological insights in COPD: an updated working disease model to broaden therapeutic vision. 30 (2021).
Gao, Z., Grout, R. W., Holtze, C., Hoffman, E. A. & Saha, P. K. A new paradigm of interactive artery/vein separation in noncontrast pulmonary CT imaging using multiscale topomorphologic opening. IEEE Trans Biomed Eng 59, 3016–3027, https://doi.org/10.1109/TBME.2012.2212894 (2012).
Chu, Y. et al. Deep learning-driven pulmonary artery and vein segmentation reveals demography-associated vasculature anatomical differences. Nat Commun 16, 2262, https://doi.org/10.1038/s41467-025-56505-6 (2025).
Katase, S. et al. Development and performance evaluation of a deep learning lung nodule detection system. BMC Med Imaging 22, 203, https://doi.org/10.1186/s12880-022-00938-8 (2022).
Sweetline, B. C., Vijayakumaran, C. & Samydurai, A. Overcoming the Challenge of Accurate Segmentation of Lung Nodules: A Multi-crop CNN Approach. J Imaging Inform Med 37, 988–1007, https://doi.org/10.1007/s10278-024-01004-1 (2024).
Zhang, H., Zhang, M., Gu, Y. & Yang, G. Z. Deep anatomy learning for lung airway and artery-vein modeling with contrast-enhanced CT synthesis. Int J Comput Assist Radiol Surg 18, 1287–1294, https://doi.org/10.1007/s11548-023-02946-7 (2023).
Cheng, H. et al. in Pattern Recognition: 27th International Conference, ICPR 2024, Kolkata, India, December 1–5, 2024, Proceedings, Part XII 422–438 (Springer-Verlag, Kolkata, India, 2024).
Rudyanto, R. D. et al. Comparing algorithms for automated vessel segmentation in computed tomography scans of the lung: the VESSEL12 study. Med Image Anal 18, 1217–1232, https://doi.org/10.1016/j.media.2014.07.003 (2014).
Zhang, M. et al. Multi-site, Multi-domain Airway Tree Modeling. Med Image Anal 90, 102957, https://doi.org/10.1016/j.media.2023.102957 (2023).
Luo, G. et al. Efficient automatic segmentation for multi-level pulmonary arteries: The PARSE challenge. arXiv:2304.03708. https://ui.adsabs.harvard.edu/abs/2023arXiv230403708L (2023).
Poletti, J. et al. Automated lung vessel segmentation reveals blood vessel volume redistribution in viral pneumonia. Eur J Radiol 150, 110259, https://doi.org/10.1016/j.ejrad.2022.110259 (2022).
Xie, K., Yang, J., Wei, D., Weng, Z. & Fua, P. Efficient anatomical labeling of pulmonary tree structures via deep point-graph representation-based implicit fields. Med Image Anal 99, 103367, https://doi.org/10.1016/j.media.2024.103367 (2025).
van Ginneken, B. A. J. Colin. LUNA16 Part 1/2, https://doi.org/10.5281/zenodo.3723295 (2019).
van Ginneken, B. A. J. Colin. LUNA16 Part 2/2, https://doi.org/10.5281/zenodo.4121926, (2019).
Fedorov, A. et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 30, 1323–1341, https://doi.org/10.1016/j.mri.2012.05.001 (2012).
Liu, J. et al. AirRC Dataset https://doi.org/10.6084/m9.figshare.26878867 (2025).
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18, 203–211, https://doi.org/10.1038/s41592-020-01008-z (2021).
Zheng, H. et al. Alleviating Class-Wise Gradient Imbalance for Pulmonary Airway Segmentation. IEEE Trans Med Imaging 40, 2452–2462, https://doi.org/10.1109/TMI.2021.3078828 (2021).
Zhang, M., Yang, G.-Z. & Gu, Y. Differentiable topology-preserved distance transform for pulmonary airway segmentation. arXiv preprint arXiv:2209.08355 (2022).
Acknowledgements
We extend our sincere gratitude to Dr. Caicun Zhou and Dr. Anwen Xiong from Shanghai East Hospital for their valuable guidance on this work.
Author information
Authors and Affiliations
Contributions
Conception and design: Jian Liu, Bing Niu, Kai Xu Administrative support: Kai Xu, Lei Wang Provision of study materials or patients: N/A Collection and assembly of data: Zheng Zhang, Bing Niu, Shuai Kang, Juan Ren, Lei Wang Data analysis and interpretation: Jian Liu, Bing Niu, Lei Wang, Zheng Zhang, Juan Ren Manuscript writing: All authors Final approval of manuscript: All authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, J., Zhang, Z., Niu, B. et al. A Custom Annotated Dataset for Segmentation of Pulmonary Veins, Arteries, and Airways. Sci Data 12, 1806 (2025). https://doi.org/10.1038/s41597-025-06074-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-06074-6