Abstract
Brain metastases (BM) are a severe complication of multiple primary malignancies and constitute the most frequent tumors of the central nervous system. Clinical imaging plays a crucial role in the management of BMs, serving as an essential tool for accurate diagnosis, effective treatment planning, and systematic follow-up care. However, progress in improving BM diagnosis and treatment is hindered by the limited availability of high-quality, annotated imaging datasets that reflect clinical variability in morphology, treatment planning, and patient characteristics. This study presents a comprehensive longitudinal dataset of 40 patients, including 744 MRI scans (baseline and follow-up), as well as 45 radiotherapy plans and 45 computed tomography scans. Detailed segmentations of 65 BMs are provided, defining three key tumor regions: enhancing tumor, edema, and necrotic core, offering a precise representation of tumor morphology for advanced quantitative analysis. This data-sharing initiative aims to support research efforts in automated BM detection, lesion segmentation, disease assessment, treatment planning, while contributing to the development of pertinent predictive and prognostic tools to enhance clinical decision-making.
Similar content being viewed by others
Background & Summary
Brain metastases (BM) are the most common type of neoplasia affecting the central nervous system, with 20–40% of cancer patients developing BM at some point during their disease progression1,2. BM are among the main complications associated with lung cancer, breast cancer and melanoma3,4,5. The advent of new therapeutic approaches is believed to have contributed to an increase in metastatic brain disease cases6,7. This is likely due to extended patient survival following diagnosis, as well as advancements in imaging technology enabling earlier and more precise detection8,9.
Management of BMs involves various therapeutic modalities: surgery, chemotherapy, immunotherapy, targeted therapies and radiotherapy10. A key challenge with radiotherapy (RT) is achieving an optimal balance between maximizing tumor control and minimizing adverse side effects. Presently, two methods of external beam radiation therapy for BMs are primarily used: whole-brain radiotherapy (WBRT) and stereotactic radiosurgery (SRS)11. WBRT is often recommended for patients with more than three BMs, as it targets the entire brain and can eliminate undetectable metastases12,13. The primary disadvantage of WBRT is that it can cause radiation-induced damage across the brain. On the other hand, SRS is typically used for patients with one to three BMs and a favorable prognosis14. In SRS, highly precise, localized radiation is applied to the BMs in a single session to optimize local tumor control while minimizing exposure to the surrounding healthy brain tissue.
Magnetic resonance imaging (MRI) is currently the modality of choice for the detection, treatment planning, and monitoring of the outcome of BM therapy. Due to its superior soft-tissue contrast and improved visualization of intracranial structures compared to other modalities, MRI is an indispensable modality in neuro-oncologic imaging15. Brain MRI protocols in neuro-oncology typically include T1-weighted (T1W), fluid-attenuated inversion recovery (FLAIR), T2-weighted (T2W) and post-contrast T1-weighted (T1-Contrast) sequences16,17. Contrast enhanced T1-weighted sequences are considered the gold standard for the detection and characterization of BMs, providing detailed information on lesion size, morphology and adjacent brain tissue17. Additionally, T2W and FLAIR images are commonly utilized to characterize BMs, as they are effective in highlighting perilesional edema typically surrounding BM lesions18.
Intracranial metastatic disease can exhibit diverse imaging patterns, with brain metastases potentially developing in the brain parenchyma, ventricles, leptomeninges, dura, bone, and extracranial soft tissues19. Precise identification of lesion number and location is crucial for surgery or radiotherapy planning. Traditionally, BMs are detected manually by a radiologist and contoured by a radiation oncologist using a radiation therapy planning software20. Automating BMs detection can serve as a valuable tool to assist clinicians in evaluating images, aiding in treatment planning for SRS, and assessing treatment outcomes during patient follow-up. Furthermore, this automation can effectively minimize human errors and accomplish tasks with minimal manual intervention. Recent advancements in deep learning have demonstrated the significant potential AI-driven methods in medical image analysis, particularly in segmentation, cancer detection and classification21,22 while some studies have reported models for BM segmentation in MRI data22,23,24,25. In addition to the automatic segmentation of BMs, morphological MRI-based features have demonstrated effectiveness in the context of other brain tumors; however, their applicability to BMs identification remains largely underexplored. Radiomic analyses have the potential to enhance the standard of care for patients with BM by extracting high-dimensional quantitative evidence that reflect imaging phenotypes from tumor segmentation, thereby supporting clinical decision-making and personalized therapy of brain cancer.
Although AI-based approaches have demonstrated encouraging outcomes in diagnosing and automatically segmenting BMs, a significant gap remains in their clinical integration and widespread adoption. A key factor contributing to this gap is their limited ability to generalize to real-world datasets. Many of these models have been developed using small, single-institution hospital datasets that lack diversity in patient demographics and imaging protocols, both of which are essential for broader clinical application26. Therefore, there is an urgent need to develop extensive, diverse and publicly accessible datasets to enhance the training of AI algorithms and to evaluate models accurately across a wide range of patient cases.
The availability of publicly accessible datasets for BMs is quite limited. The Cancer Imaging Archive27 remains the most prominent repository for cancer imaging data – it offers data for various human cancers, including brain cancers. Currently, dedicated publicly available BM datasets28,29,30, as well as the BraTS-METS 202323 dataset, are limited in that they only include pre-treatment segmentations and MRI data, capturing only a limited range of tumor subregions – typically the contrast-enhancing region of the tumor or the necrotic tumor area28,30. Moreover, existing studies provide minimal to no information about the therapeutic approaches BM patients undergone, including radiation, chemotherapy, or immunotherapy. Addressing these gaps is crucial for advancing AI-driven BM analysis and improving clinical decision-making.
In this contribution, we present a unique BM dataset of cancer patients in Cyprus, which includes standard MRI sequences (T1W, T2W, T1-Contrast, FLAIR; see Fig. 1), the radiotherapy plan (RTP) and computed tomography (CT) scans, and detailed annotations of tumor subregions – specifically labels for the enhancing tumor, the edema, the necrotic region. Additionally, we provide two supplementary Excel files containing clinical evidence: one file includes essential demographic, clinical, and treatment-related information, offering valuable insights for medical analysis and AI-driven research, while the other file provides quantitative imaging data for each patient case.
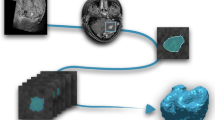
Representative MRI scans of a patient displaying the brain in four imaging modalities: (a) T1-weighted, (b) T2-weighted, (c) post-contrast T1-weighted (T1-Contrast), and (d) fluid-attenuated inversion recovery (FLAIR). (e) Tumor subregions are delineated according to the BraTS annotation standards, with necrotic core (red), peritumoral edema (green), and enhancing tumor (yellow). (f) The radiotherapy overlaid on a computed tomography scan of brain tumor, illustrating dose distribution and anatomical structures involved in treatment.
Each patient record includes pre-treatment tumor segmentations and MRI scans, as well as post-treatment follow-up imaging data. All patients had at least one follow-up, conducted at predetermined intervals of 6 weeks, 3 months, 6 months, 9 months and 12 months, enabling a comprehensive longitudinal assessment of treatment response and disease progression. Importantly, the inclusion of post-treatment imaging data addresses a core requirement of the BraTS-METS 2025 Lighthouse Challenge and enhances the dataset’s utility for developing and validating AI algorithms.
The present data resource is designed to support the training and validation of future machine learning and in silico models for BM diagnosis and therapeutic monitoring, with the ultimate goal of facilitating their translation and adoption in clinical practice. The multiple follow-up scans (at predefined time points) that are available in the present dataset further enables the investigation of longitudinal disease dynamics, treatment response, and temporal evolution of imaging biomarkers. This distinguishes our dataset from existing BM resources and provides a foundation for developing models that account for time-dependent changes in tumor biology.
Methods
Subject characteristic
The data collected in this study include the pretreatment and follow-up studies from 6 different diagnostic centers in Cyprus, where all patients were identified from a reference clinical oncology center, the Bank of Cyprus Oncology Centre (BoCOC) between 2019 and 2024. All recruited patients signed informed consent. Inclusion required a clinical or pathological diagnosis of BM and the availability of all four standard MRI sequences (i.e., T1W, T2W, T1-Contrast, and FLAIR). All scans were qualitatively inspected for image quality by an experienced radiologist, and those exhibiting significant motion-related blurring or ghosting artifacts were excluded. Additionally, quantitative consistency of the imaging data was assessed at the protocol level by reviewing voxel-related parameters, including slice thickness and slice spacing, extracted from DICOM headers and are summarized in Table 1. The dataset comprises of a total of 40 patients. The primary tumor origins among these patients were as follows: non-small cell lung cancer (65%), breast cancer (32.5%), and small-cell lung cancer (2.5%). In summary, the cohort of 40 patients included in this study exhibit a total of 65 BMs. The dataset consists of 40 patients, including 744 distinct MRI scans, along with 45 unique RTP and 45 CT scans. Notably, 5 patients required two separate RTP and CT due to the emergence of new brain metastases. All patients underwent stereotactic radiosurgery or stereotactic fractionated radiotherapy. Lesions measuring less than 3 millimeters in size were ignored and excluded from imaging analysis to ensure accurate and reliable assessments.
Ethical approval
The present study was conducted according to the guidelines of the Declaration of Helsinki and has been approved by the Cyprus National Bioethics Committee (EEBK/EΠ/2021/72; February 24, 2022). All participants provided their written consent for data anonymization and approved open publication of the data.
Clinical data and anonymization
Clinical data were retrieved from the BoCOC electronic medical record system and include the following information: ethnicity, gender, age at diagnosis, the origin of the primary tumor, the treatment details related to chemotherapy and radiosurgery, the Karnofsky performance status, the neurocognitive status, presence and extent of extracranial disease, and the time to death or last documented entry in the electronic medical record as of December 2024. For each metastatic lesion, the recorded details included a unique identifier to differentiate it from other metastases within the same patient, its precise anatomical location within the brain (classified as hemispheric frontal, temporal, parietal, or occipital, brainstem, or cerebellar, including laterality), the date of initial detection on MRI, and the corresponding treatment details administered. Treatment-related data included the type of therapy dosages, number of SRT fractions, start dates, and where available, end dates of treatment.
All DICOM files were de-identified prior to deposition using the anonymization tools integrated in the Eclipse Radiotherapy Treatment Planning System (Varian Medical Systems). The de-identification process was executed on site at the BoCOC research data server immediately following receipt of the DICOM images from the PACS production system. This process automatically removed protected health information, private DICOM tags, and any fields containing sensitive or identifying data, ensuring participant confidentiality.
Image acquisition
Table 1 presents a summary of all imaging parameters for the MRI acquisitions. T1W contrast-enhanced images were acquired following intravenous administration of a paramagnetic contrast agent (Gadovist) at a dose of approximately 0.1 mmol/kg body weight. The 185 MRI studies were conducted under free-breathing conditions using either 3 T (n = 126; 68.5%) or 1.5 T (n = 58; 31.5%) MRI scanners. The MRI examinations were performed using scanners manufactured by Philips (n = 126; 68.5%), Siemens (n = 55; 29.9%) and Toshiba (n = 3; 1.6%).
Image preprocessing
The imaging data, initially available in the Digital Imaging and Communications in Medicine (DICOM) format, were converted to the Neuroimaging Informatics Technology Initiative (NIfTI) format using dcm2niix31. This conversion ensured compatibility with the computational tools used while maintaining image quality, spatial orientation and resolution. Subsequently, all MRI data were transformed to the Brain Tumor Segmentation (BraTS) spatial framework using the BraTS-Preprocessor32, to adhere to the requirements of the BraTS challenge. This transformation process involved resampling and registering all images to the BraTS-defined space, thereby achieving uniformity in voxel dimensions and anatomical alignment. By adhering to the standards BraTS challenge, the dataset was optimized for interoperability, usability and reproducibility, thus, enabling seamless application in prospective BraTS challenges and promoting the development of brain tumor segmentation algorithms. Lastly, the RTP and CT data were aligned to the processed T1W images using non-rigid registration33, while ensuring precise spatial correspondence with the BraTS standards to facilitate further utilization of the data.
Segmentation procedure
The MRI scans, were processed using multiple segmentation methodologies, including the deep learning model DeepBraTumIA34, the nnU-Net35 model trained on the BraTS 2020 dataset, and the proprietary software Imalytics Preclinical36. Both DeepBraTumIA and nnU-Net models were used to automatically segment each MRI scan into three tumor subregions: enhancing tumor edema (T2/FLAIR hyperintensity) and tumor necrosis (see Fig. 1). In cases where patients had multiple metastases, DeepBraTumIA and nnU-Net were unable to detect all lesions. To address this limitation, a semi-automated segmentation method using Imalytics Preclinical36, was carried out. In this approach, tumors were initially manually delineated across several central slices of the 3D volume, followed by automatic detection across all slices using built-in algorithms of the software. The edema and necrotic tissue segmentations were performed manually by experienced professionals to ensure accuracy. In addition, the segmentation of white matter, grey matter, ventricle and cerebrospinal fluid components of normal brain tissue were automatically annotated using a geodesical information flows-based algorithm36 that is incorporated into the open-source toolkit NiftySeg. The segmentation outputs generated by DeepBraTumIA, nnU-Net and Imalytics Preclinical, were subsequently transformed into the BraTS space (Fig. 1e) to ensure accurate and reliable alignment with the corresponding MRIs. To guarantee consistency and clinical reliability, all segmentations were thoroughly reviewed and manually refined by a certified neuroradiologist (L.S.) with over 10 years of clinical experience. Open-source software ITK-SNAP 4.0.2 was used to visualize and verify the segmentation results during the revision process.
Radiomic-based analysis
A comprehensive radiomic-based analysis pipeline was conducted using all available MRI modalities and corresponding features were extracted from the brain region with tissue necrosis, the tumor region and the edema, respectively. Prior to the radiomics feature extraction, N4 bias field correction was utilized using the SimpleITK Python package (version 2.4.1) to surpass bias field degradation in the acquired MRIs, and to increase the number of reproducible features37,38. Subsequently, image slices were subjected to intensity clipping using the 0.1 and 99.9 percentile of the 1-dimensional histogram as thresholds to reduce outlier signal intensities (supported by function ClampImageFilter of the SimpleITK Python package). Image rescaling (0 to 1024) was then performed (rescale_intensity from Python package scikit-image, version 0.25) and pre-processed images were imported into the Pyradiomics Python package (version 3.1.0)39, deriving a total of 110 features from each MRI modality, segmentation region, and acquisition time. These features consist of 19 first-order statistics, 16 3D shape-based descriptors, and 75 textural characteristics: 24 from the gray level co-occurrence matrix, 16 from the Gray Level Run Length matrix, 5 from the neighboring Gray Tone Difference matrix, 16 Gray Level Size Zone features, and 14 from the Gray Level Dependence matrix. Since all images were isotropic interpolated during the segmentation phase, “interpolator” from the Pyradiomics was disabled. Discretization of the image gray level was set to a bin width of 5.
Data Record
The dataset has been deposited in Zenodo40. The present dataset contains medical imaging data (MRI scans: baseline and follow-up timepoints; RTP scans; CT scans) and two Excel spreadsheets that contain the clinical and demographic data (‘PROTEAS-Clinical_and_demographic_data.xlsx’) and the radiomics data (‘PROTEAS-MRI_radiomics_data.xlsx’) from all 40 patients included in this study. For each patient, raw pretreatment and follow-up scans are stored in DICOM file format, which preserves the original medical image coordinates. Tumor segmentations from baseline and follow-up scans are provided in NIfTI format after preprocessing, as described in the Image Preprocessing section. All patients are assigned a unique generic identifier; for example, the data for patient 01 are stored in folder ‘P01’, for patient 11 in ‘P11’, and so on. An exception applies to patients 04, 07 17, 20 and 23, for whom two directories are provided (e.g., in patient 23 we have folders: ‘P23a’ to ‘P23b’). As reported in the clinical data spreadsheet, these patients underwent two rounds of radiotherapy due to the presence of distinct BM located in separate regions of the brain. Each patient directory contains a folder entitled ‘BraTS’, which includes subfolders labeled ‘baseline,’ ‘fu1,’ ‘fu2’ and so forth, depending on the number of available follow-up scans. Each of these subfolders contains four NifTI files, provided in BraTS space, corresponding to the MRI sequences collected in this study: ‘t1.nii.gz’ (T1W), ‘t2.nii.gz’ (T2W), ‘t1c.nii.gz’ (T1-Contrast), and ‘fla.nii.gz’ (FLAIR). In addition, each patient directory contains: (i) a NifTI file named ‘P??_brain_mask.nii.gz’ with the healthy brain tissue regions, (ii) a NifTI file named ‘P??_CT.nii.gz’ with the CT data, (iii) a NifTI file named ‘P??_RTP.nii.gz’ file with the RTP data, and (iv) a folder entitled ‘tumor_segmentation’ that includes the tumor/edema/necrosis masks for the baseline scan (named ‘P??_tumor_mask_baseline.nii.gz’) and for each available follow-up respectively (e.g., the 2nd follow-up for patient 30 the filename is ‘P30_tumor_mask_fu2.nii.gz’). All brain and tumor segmentation files adhere to a consistent label convention: ‘Label 1’ represents the necrotic core, ‘Label 2’ represents the enhancing tumor region, ‘Label 3’ represents the edema, ‘Label 10’ represents the brain ventricles, ‘Label 30’ represents the white matter, ‘Label 40’ represents the grey matter, and ‘Label 50’ represents the cerebrospinal fluid region.
In summary, the present dataset provides pretreatment MRI scans and multiple follow-up time points, along with corresponding tumor segmentations and radiomic features for each patient. The dataset enables the investigation of disease progression, treatment response, and temporal changes in tumor subregions (necrotic core, enhancing tumor, edema). Researchers can also study the evolution of imaging biomarkers and radiomic signatures over time, therefore, making the present dataset a valuable resource for longitudinal analyses and predictive modeling in brain metastasis research.
Technical Validation
The study included only patients with a confirmed BM, with primary tumors pathologically and clinically confirmed by a radiation oncologist (M.T.) prior to inclusion. Data curation and evaluation of inclusion criteria were conducted by an MRI analysis professional (D.F.). The final dataset comprised only patients with high-quality T1W, T2W, T1-Contrast and FLAIR images, free from significant motion artifacts. Lastly, all automatic and semi-automatic segmentations performed in this study were manually corrected by an expert neuroradiologist (L.S.) after being carried out by medical image analysis experts (D.F., C.P.P).
Usage Notes
To process the medical images and segmentations provided in this work, it is strongly advised to use a medical imaging software that supports either the DICOM or NIfTI format that ensures consistent handling of the images’ physical space and orientation. We confirmed that all NIfTI files, including segmentations and images, were successfully loaded using FSLeyes (version 1.12.6), ITK-SNAP 4.2.2 and 3D Slicer (version 5.8.1) software, and DICOM files were successfully loaded using Horos (version 4.01) and 3D Slicer (version 5.8.1) software.
Data availability
The dataset can be downloaded online directly from the repository: https://doi.org/10.5281/zenodo.17253793.
Code availability
The Python code used for feature extraction is freely available on GitHub: https://github.com/InSilicoModellingGroup/pyRadiomicBM. Proper documentation of the code is provided therein and a comprehensive README.md file is available for how to run the algorithms, reproduce the results of this manuscript, and facilitate customization of the radiomics analysis.
References
Hügel, M. et al. Long-term survival in patients with brain metastases—clinical characterization of a rare scenario. Strahlenther. Onkol. 200, 335–345 (2024).
Mitchell, D. K. et al. Brain metastases: an update on the multi-disciplinary approach of clinical management. Neurochirurgie 68, 69–85 (2022).
Kotecha, R., Gondi, V., Ahluwalia, M. S., Brastianos, P. K. & Mehta, M. P. Recent advances in managing brain metastasis. F1000Res 7, F1000 Faculty Rev–1772 (2018).
Achrol, A. S. et al. Brain metastases. Nat. Rev. Dis. Primers 5, 5 (2019).
Lekkala, M. R. & Mullangi, S. Malignant melanoma metastatic to the central nervous system. In StatPearls (StatPearls Publishing, 2023).
DiLuna, M. L., King, J. T. Jr, Knisely, J. P. & Chiang, V. L. Prognostic factors for survival after stereotactic radiosurgery vary with the number of cerebral metastases. Cancer 109, 135–145 (2007).
Nayak, L., Lee, E. Q. & Wen, P. Y. Epidemiology of brain metastases. Curr. Oncol. Rep. 14, 48–54 (2012).
Tong, E., McCullagh, K. L. & Lv, M. Advanced imaging of brain metastases: from augmenting visualization and improving diagnosis to evaluating treatment response. Front. Neurol. 11, 1–14 (2020).
Lignelli, A. & Khandji, A. G. Review of imaging techniques in the diagnosis and management of brain metastases. Neurosurg. Clin. N. Am. 22, 15–25 (2011).
Eichler, A. F. & Loeffler, J. S. Multidisciplinary management of brain metastases. Oncologist 12, 884–898 (2007).
van Grinsven, E. E., Nagtegaal, S. H. J., Verhoeff, J. J. C. & van Zandvoort, M. J. E. The impact of stereotactic or whole brain radiotherapy on neurocognitive functioning in adult patients with brain metastases: a systematic review and meta-analysis. Oncol. Res. Treat. 44, 622–636 (2021).
Khuntia, D., Brown, P., Li, J. & Mehta, M. P. Whole-brain radiotherapy in the management of brain metastasis. J. Clin. Oncol. 24, 1295–1304 (2006).
Mehta, M. P. The controversy surrounding the use of whole-brain radiotherapy in brain metastases patients. Neuro Oncol. 17, 919–923 (2015).
Pinkham, M. B., Whitfield, G. A. & Brada, M. New developments in intracranial stereotactic radiotherapy for metastases. Clin. Oncol. (R. Coll. Radiol.) 27, 316–323 (2015).
Brindle, K. M., Izquierdo-García, J. L., Lewis, D. Y., Mair, R. J. & Wright, A. J. Brain tumor imaging. J. Clin. Oncol. 35, 2432–2438 (2017).
Kaufmann, T. J. et al. Consensus recommendations for a standardized brain tumor imaging protocol for clinical trials in brain metastases. Neuro Oncol. 22, 757–772 (2020).
Alibek, S. et al. Contrast-enhanced T1-weighted fluid-attenuated inversion-recovery BLADE magnetic resonance imaging of the brain: an alternative to spin-echo technique for detection of brain lesions in the unsedated pediatric patient? Acad. Radiol. 15, 986–995 (2008).
Derks, S. H. A. E., van der Veldt, A. A. M. & Smits, M. Brain metastases: the role of clinical imaging. Br. J. Radiol. 95, 20210944 (2022).
Barnholtz-Sloan, J. S. et al. Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J. Clin. Oncol. 22, 2865–2872 (2004).
Aizer, A. A. et al. Brain metastases: a Society for Neuro-Oncology (SNO) consensus review on current management and future directions. Neuro Oncol. 24, 1613–1646 (2022).
Conson, M. et al. Automated delineation of brain structures in patients undergoing radiotherapy for primary brain tumors: from atlas to dose-volume histograms. Radiother. Oncol. 112, 326–331 (2014).
Pennig, L. et al. Automated detection and segmentation of brain metastases in malignant melanoma: evaluation of a dedicated deep learning model. AJNR Am. J. Neuroradiol. 42, 655–662 (2021).
Moawad, A. W. et al. The Brain Tumor Segmentation - Metastases (BraTS-METS) Challenge 2023: brain metastasis segmentation on pre-treatment MRI. arXiv preprint arXiv:2306.00838v3 (2024).
Liu, Y. et al. A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS One 12, e0185844 (2017).
Charron, O. et al. Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput. Biol. Med. 95, 43–54 (2018).
Ghaderi, S. et al. A systematic review of brain metastases from lung cancer using magnetic resonance neuroimaging: clinical and technical aspects. J. Med. Radiat. Sci. 71, 269–289 (2024).
Clark, K. et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057 (2013).
Ocaña-Tienda, B. et al. A comprehensive dataset of annotated brain metastasis MR images with clinical and radiomic data. Sci. Data 10, 208 (2023).
BrainMetShare, Center for Artificial Intelligence in Medicine & Imaging. https://doi.org/10.71718/z66c-qr59 (2019).
Ramakrishnan, D. et al. A large open access dataset of brain metastasis 3D segmentations on MRI with clinical and imaging information. Sci. Data 11, 254 (2024).
Li, X., Morgan, P. S., Ashburner, J., Smith, J. & Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 264, 47–56 (2016).
Kofler, F. et al. BraTS Toolkit: translating BraTS brain tumor segmentation algorithms into clinical and scientific practice. Front. Neurosci. 14, 125 (2020).
Modat, M. et al. Fast free-form deformation using graphics processing units. Comput. Methods Programs Biomed. 98, 278–284 (2010).
German Cancer Research Center. DeepBraTumIA: Deep learning-based brain tumor segmentation tool. NeuroImaging Tools and Resources Collaboratory (NITRC) https://www.nitrc.org/projects/deepbratumia (2021).
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Gremse, F. et al. Imalytics Preclinical: interactive analysis of biomedical volume data. Theranostics 6, 328–341 (2016).
Cardoso, M. J. et al. Geodesic information flows: spatially-variant graphs and their application to segmentation and fusion. IEEE Trans. Med. Imaging 34, 1976–1988 (2015).
Stamoulou, E. et al. Harmonization strategies in multicenter MRI-based radiomics. J. Imaging 8, 303 (2022).
van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104–e107 (2017).
Flouri, D. et al. Diagnostic and follow-up MRIs, CTs, radiotherapy and radiomics data of brain metastases. Zenodo https://doi.org/10.5281/zenodo.15499111 (2025).
Acknowledgements
This work was financially supported by the Cyprus Cancer Research Institute through the PROTEAS project (Call: Bridges in Research Excellence CCRI_2020_FUN_001; Grant ID: CCRI_2021_FA_LE_105). D.F. is a recipient of a Marie Skłodowska-Curie Actions postdoctoral fellowship for the InSilicoPlacenta project (grant agreement ID: 101108945). G.C.M. is a recipient of a Marie Skłodowska-Curie Actions postdoctoral fellowship for the GRANITE project (grant agreement ID: 101153374). V.V. acknowledges the financial support of the EIC Pathfinder Open project SoftReach (grant agreement ID: 101099145). The authors are grateful to Dr Felix Gremse for his technical support using the Imalytics Preclinical software.
Author information
Authors and Affiliations
Contributions
D.F. – data export and quality control, brain segmentations, tumor segmentations, dataset assembly, dataset publication, manuscript preparation. C.P.P. – tumor segmentations, dataset assembly, manuscript revision. G.M. – radiomics feature analysis, manuscript revision. E.I. – manuscript revision. G.K. – clinical data collection, image transfer and de-identification, manuscript revision. E.P. – clinical data collection, manuscript revision. A.C. – conceptualization, manuscript revision. L.S. – conceptualization, imaging data collection and evaluation, tumor segmentations, manuscript revision. M.T. – clinical data collection, manuscript revision. V.V. – conceptualization, project supervisor, dataset publication, funding acquisition, manuscript revision.
Corresponding authors
Ethics declarations
Competing interests
L.S. collaborates with the Ayios Therissos Diagnostic Center (https://ayiostherissos.eu/en/), a contributing site to the PROTEAS study. This collaboration did not influence the design, data analysis, or interpretation of this study, and does not constitute a competing interest. The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Flouri, D., Papanikas, CP., Manikis, G.C. et al. A longitudinal MRI dataset of brain metastases with tumor segmentations, clinical & radiomic data. Sci Data 12, 1828 (2025). https://doi.org/10.1038/s41597-025-06131-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-06131-0