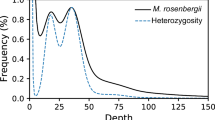
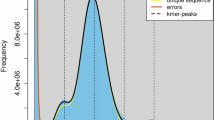
Abstract
The giant pangasius (Pangasius sanitwongsei) is a critically endangered freshwater species with considerable ecological and economic significance in Southeast Asia and southern China. However, the lack of a high-quality reference genome has limited studies on its genetic adaptation and conservation strategies. Here, we present a chromosome-scale genome assembly of P. sanitwongsei, generated using PacBio HiFi long-read sequencing and Hi-C chromatin conformation capture. The final assembly spans 808.85 Mb, with a contig N50 of 18.70 Mb and a scaffold N50 of 28.10 Mb, achieving a BUSCO gene completeness of 99.15%. A total of 23,469 protein-coding genes were annotated, and 96.97% of the genes were functionally characterized. This high-quality genomic resource provides crucial insights into the adaptive evolution of Pangasiidae catfishes and offers a valuable foundation for conservation genomics, adaptive evolution, population genetics, and sustainable aquaculture development.
Similar content being viewed by others
Data availability
The raw sequencing datasets have been deposited in the NCBI Sequence Read Archive under accession SRP57947851. The assembled P. sanitwongsei genome is available in GenBank under accession GCA_051225755.152, and the corresponding genome annotation files has been archived on Figshare (https://doi.org/10.6084/m9.figshare.29715215)53.
Code availability
No special codes or scripts were used in this work, and data processing was carried out based on the protocols and manuals of the corresponding bioinformatics software.
References
Froese, R. & Pauly, D. (Fisheries Centre, University of British Columbia Los Baños, Philippines, 2010).
Kang, B. & Huang, X. Mekong fishes: Biogeography, migration, resources, threats, and conservation. Rev Fish Sci Aquac 30, 170–194 (2022).
Roberts, T. R. & Vidthayanon, C. Systematic revision of the Asian catfish family Pangasiidae, with biological observations and descriptions of three new species. Proceedings of the Academy of Natural Sciences of Philadelphia, 97-143 (1991).
Roberts, T. R. & Baird, I. G. Traditional fisheries and fish ecology on the Mekong River at Khone Waterfalls in southern Laos. Natural History Bulletin of the Siam Society 43, 219–262 (1995).
Chanthasoo, M., Wiwatcharakoset, S. & Lisanga, S. Breeding of Chao Phaya Giant catfish (Pangasius Sanitwongsei) (1990).
Sutthi, N., Panase, A., Phinrub, W., Srisuttha, P. & Panase, P. Cold shock and its effect on biochemical indices, cortisol and electrolyte changes in Chao Phraya catfish, Pangasius sanitwongsei smith, 1931. Comparative Clinical Pathology 31, 757–764 (2022).
Na-Nakorn, U. et al. Genetic Diversity of the Vulnerable Pangasius sanitwongsei using Microsatellite DNA and 16S rRNA. Journal of Fisheries and Environment 33, 24–40 (2009).
Makinen, T., Weyl, O. L. F., Van der Walt, K.-A. & Swartz, E. R. First record of an introduction of the giant pangasius, Pangasius sanitwongsei Smith 1931, into an African river. African Zoology 48, 388–391 (2013).
Hogan, Z., Na-Nakorn, U. & Kong, H. Threatened fishes of the world: Pangasius sanitwongsei Smith 1931 (Siluriformes: Pangasiidae). Environmental Biology of Fishes 84, 305–306 (2009).
Campbell, T., Pin, K., Ngor, P. B. & Hogan, Z. Conserving Mekong megafishes: Current status and critical threats in Cambodia. Water 12, 1820 (2020).
Baird, I. G. & Hogan, Z. S. Hydropower Dam development and fish biodiversity in the Mekong River Basin: A review. Water 15, 1352 (2023).
Jutagate, T. & Rattanachai, A. Inland fishery resource enhancement and conservation in Thailand. Inland fisheries resource enhancement and conservation in Asia 133 (2010).
Kitcharoen, N., Nakkham, K. & Mengumphan, K. A study on growth performance of interspecific crosses-hybrid cat fish spices: Buk Siam hybrid catfish (male Pangasianodon gigas x female P. hypophthalmus) Pangosius larnaudii and Pangasius sanitwongsei (2022).
Karinthanyakit, W. & Jondeung, A. Molecular phylogenetic relationships of pangasiid and schilbid catfishes in Thailand. J Fish Biol 80, 2549–2570 (2012).
Duong, T. Y. et al. Mitophylogeny of Pangasiid catfishes and its taxonomic implications for Pangasiidae and the suborder Siluroidei. Zoological studies 62, e48 (2023).
Na‐Nakorn, U., Sriphairoj, K., Sukmanomon, S., Poompuang, S. & Kamonrat, W. Polymorphic microsatellite primers developed from DNA of the endangered Mekong giant catfish, Pangasianodon gigas (Chevey) and cross‐species amplification in three species of Pangasius. Molecular Ecology Notes 6, 1174–1176 (2006).
Wei, L. et al. Complete mitochondrial genome and phylogenetic position of Pangasius sanitwongsei (Siluriformes: Pangasiidae). Mitochondrial DNA Part B 5, 945–946 (2020).
Sriphairoj, K., Na-Nakorn, U. & Klinbunga, S. Species identification of non-hybrid and hybrid Pangasiid catfish using polymerase chain reaction-restriction fragment length polymorphism. Agriculture and Natural Resources 52, 99–105 (2018).
Gao, Z. et al. A chromosome-level genome assembly of the striped catfish (Pangasianodon hypophthalmus). Genomics 113, 3349–3356 (2021).
Hai, D. M. et al. A high-quality genome assembly of striped catfish (pangasianodon hypophthalmus) based on highly accurate long-read hifi sequencing data. Genes 13, 923 (2022).
Kim, O. T. P. et al. A draft genome of the striped catfish, Pangasianodon hypophthalmus, for comparative analysis of genes relevant to development and a resource for aquaculture improvement. Bmc Genomics 19, 733 (2018).
Wen, M. et al. An ancient truncated duplication of the anti‐Müllerian hormone receptor type 2 gene is a potential conserved master sex determinant in the Pangasiidae catfish family. Mol Ecol Resour 22, 2411–2428 (2022).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Ranallo-Benavidez, T. R., Jaron, K. S. & Schatz, M. C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun 11, 1432 (2020).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods 18, 170–175 (2021).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell systems 3, 95–98 (2016).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. bioinformatics 25, 1754–1760 (2009).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell systems 3, 99–101 (2016).
Wang, X. & Wang, L. GMATA: an integrated software package for genome-scale SSR mining, marker development and viewing. Frontiers in plant science 7, 215951 (2016).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27, 573–580 (1999).
Han, Y. & Wessler, S. R. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res 38, e199–e199 (2010).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proceedings of the National Academy of Sciences 117, 9451–9457 (2020).
Abrusán, G., Grundmann, N., DeMester, L. & Makalowski, W. TEclass—a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25, 1329–1330 (2009).
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenetic and genome research 110, 462–467 (2005).
Hubley, R. et al. The Dfam database of repetitive DNA families. Nucleic Acids Res 44, D81–D89 (2016).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology 37, 907–915 (2019).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature biotechnology 33, 290–295 (2015).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature protocols 8, 1494–1512 (2013).
Li, H. Protein-to-genome alignment with miniprot. Bioinformatics 39, btad014 (2023).
Brůna, T., Hoff, K. J., Lomsadze, A., Stanke, M. & Borodovsky, M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR genomics and bioinformatics 3, lqaa108 (2021).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34, W435–W439 (2006).
Bruna, T., Lomsadze, A. & Borodovsky, M. GeneMark-ETP: automatic gene finding in eukaryotic genomes in consistency with extrinsic data. BioRxiv, 2023-2001 (2023).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome biology 9, 1–22 (2008).
Buchfink, B., Reuter, K. & Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18, 366–368 (2021).
Bairoch, A. et al. The universal protein resource (UniProt). Nucleic Acids Res 33, D154–D159 (2005).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000).
Tatusov, R. L. et al. The COG database: an updated version includes eukaryotes. BMC bioinformatics 4, 1–14 (2003).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat Genet 25, 25–29 (2000).
Kanz, C. et al. The EMBL nucleotide sequence database. Nucleic Acids Res 33, D29–D33 (2005).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP579478 (2025).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_051225755.1 (2025).
Liu, H. Chromosome-level genome assembly of the Siniperca obscura. Figshare https://doi.org/10.6084/m9.figshare.29715215 (2025).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Tang, H. et al. Synteny and collinearity in plant genomes. Science 320, 486–488 (2008).
Tang, H. et al. JCVI: A versatile toolkit for comparative genomics analysis. iMeta, e211 (2024).
Marçais, G. et al. MUMmer4: A fast and versatile genome alignment system. PLoS computational biology 14, e1005944 (2018).
Goel, M., Sun, H., Jiao, W.-B. & Schneeberger, K. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome biology 20, 277 (2019).
Acknowledgements
We acknowledge financial support from the Guangxi Key R&D Program Agriculture and Rural Areas (AB2506910048), National Modern Agriculture Industry Technology System Special Project (CARS-46), Central Public-interest Scientific Institution Basal Research Fund, CAFS (2025XK01, 2025SJHX1, 2023TD37), China-ASEAN Maritime Cooperation Fund (CAMC-2018F), Guangdong Province Rural Revitalization Strategy Special Fund (2023-SJS-00-001).
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gan, B., Wei, L., Ma, Y. et al. A chromosome-level reference genome assembly of the giant pangasius (Pangasius sanitwongsei). Sci Data (2025). https://doi.org/10.1038/s41597-025-06445-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-06445-z