Abstract
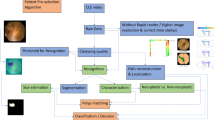
High-quality segmentation datasets are essential for advancing AI applications in medical imaging. However, it is challenging to generate such datasets for highly variable and complex organs like the colon. We introduce a dataset of 435 human colons, segmented from Computed Tomography Colonography (CTC) obtained from the publicly available The Cancer Imaging Archive (TCIA). Each scan includes a mask of the whole colon, including collapsed segments and the fluid, and a mask of only the gas-filled parts of the colon. The colon segmentation accuracy has been clinically validated by an expert abdominal radiologist. This is the first open-access dataset of segmented colons derived from CTC. This resource enables population-scale radiologic studies, supports the development of AI-based image analysis tools, and facilitates the creation of anatomically accurate digital models and simulators, both virtual and physical.
Similar content being viewed by others
Data availability
The dataset is hosted in the Open Science Framework (OSF) repository “HQColon: High-Resolution Human Colon Segmentation” (https://doi.org/10.17605/OSF.IO/8TKPM).
Code availability
The code for the semi-automatic segmentation of the colon can be found in the following GitHub repository https://github.com/horizon-europe-2023-ire/colon-segmentation-dataset. The RootPainter project for segmenting the colon fluid is available in the folder “root_painter_colon_fluid_project.zip” of the OSF repository11.
References
Alabduljabbar, A., Khan, S. U., Alsuhaibani, A., Almarshad, F. & Altherwy, Y. N. Medical imaging datasets, preparation, and availability for artificial intelligence in medical imaging. Journal of Alzheimer’s Disease Reports 8, 1471–1483 (2024).
Liu, X. et al. Towards more precise automatic analysis: a systematic review of deep learning-based multi-organ segmentation. BioMedical Engineering OnLine 23, 52 (2024).
Starck, S. et al. Using uk biobank data to establish population-specific atlases from whole body mri. Communications Medicine 4, 237 (2024).
Tang, C. et al. A roadmap for the development of human body digital twins. Nature Reviews Electrical Engineering 1(3), 199–207 (2024).
Finocchiaro, M. et al. Training simulators for gastrointestinal endoscopy: Current and future perspectives. Cancers 13(6), 1427 (2021).
Pore, A. et al. Colonoscopy navigation using end-to-end deep visuomotor control: A user study. In: 2022 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS). pp. 9582–9588. IEEE (2022).
Mang, T., Graser, A., Schima, W. & Maier, A. Ct colonography: techniques, indications, findings. European journal of Radiology 61(3), 388–399 (2007).
Smith, K. et al. Data from ct colonography https://doi.org/10.7937/K9/TCIA.2015.NWTESAY1 (2015).
Smith, A. G. et al. Rootpainter: deep learning segmentation of biological images with corrective annotation. New Phytologist 236(2), 774–791 (2022).
Wasserthal, J. et al. Totalsegmentator: robust segmentation of 104 anatomic structures in ct images. Radiology: Artificial Intelligence 5(5) (2023).
Finocchiaro, M., Stern, R., Ganz, M. High-resolution human colon segmentation dataset, Open Science Framework, https://doi.org/10.17605/OSF.IO/8TKPM (2025).
Acknowledgements
This work is funded by the European Union, grant number 101135082. Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Health and Digital Executive Agency. Neither the European Union nor the granting authority can be held responsible for them. Abraham George Smith is funded by Novo Nordisk Foundation Grant [NNF22OC0080177].
Author information
Authors and Affiliations
Contributions
Martina Finocchiaro designed and implemented the segmentation methods with contributions from Ronja Stern and Abraham George Smith; performed post-processing, quality checks and manual corrections of the segmentatios; designed the clinical validation protocol with input from Kristoffer Cold and Lars Konge; designed and developed the interface for clinical validation; analyzed the results and wrote the manuscript. Martina Finocchiaro and Ronja Stern created the data and code repositories. Rikke Vilhelmsborg clinically validated the segmentations. Kenny Erleben advised on the technical content of the research project. Melanie Ganz supervised and coordinated the research project. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Finocchiaro, M., Stern, R., Vilhelmsborg, R. et al. Clinically validated dataset of 435 human colons segmented from CT colonography. Sci Data (2026). https://doi.org/10.1038/s41597-025-06518-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-06518-z