Abstract
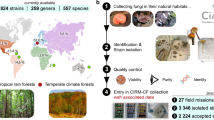
The fungal kingdom represents a greatly untapped resource to produce a wide range of bioactive secondary metabolites, including antibiotics, anticancer agents, industrially significant dyes and enzymes. To-date, it is estimated only less than 5% of all fungi have been characterised, a deficit that is especially pronounced in tropical regions like Singapore, where fungal diversity remains underexplored compared to northern hemisphere counterparts. This underlines the urgency and importance of our research which motivated the creation of our curated dataset, aiming to address this gap and contribute to understanding the broader ecosystem. We developed a generalisable cultivation workflow that enables systematic strain preparation, supports high-resolution imaging, and yields sufficient fungal biomass amenable for genomic analyses. This resulted in a diverse collection of 518 phylogenetically and ecologically varied fungal strains from both terrestrial and marine environments in biodiverse Singapore. The curated dataset from this project captures both taxonomic identity and colony-level morphological traits serving as a foundation for visual phenotype to taxonomy mapping through the integration of computer vision.
Similar content being viewed by others
Data availability
All dataset components have been made publicly available on Figshare33. The 18S sequences have also been deposited to the NCBI SRA under the accession number SRP647581 (https://identifiers.org/ncbi/insdc.sra:SRP647581)34.
Code availability
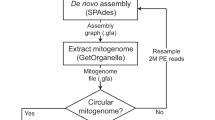
Scripts for (1) 18S rRNA prediction with Barrnap, (2) MAFFT alignment, (3) FastTree maximum-likelihood tree inference, (4) consensus tree construction and (5) ResNet-50 embedding generation are available on the GitHub repository (https://github.com/twxdarren/fungal-trove.git) and Figshare33.
References
Case, N. T. et al. The future of fungi: threats and opportunities. G3 Genes|Genomes|Genetics 12, jkac224 (2022).
Peay, K. G., Kennedy, P. G. & Talbot, J. M. Dimensions of biodiversity in the Earth mycobiome. Nat Rev Microbiol 14, 434–447 (2016).
Tedersoo, L. et al. Global diversity and geography of soil fungi. Science 346, 1256688 (2014).
Bhattarai, K., Bhattarai, K., Kabir, M. E., Bastola, R. & Baral, B. Fungal natural products galaxy: Biochemistry and molecular genetics toward blockbuster drugs discovery. in Advances in Genetics vol. 107 193–284 (Elsevier, 2021).
Danner, C., Mach, R. L. & Mach-Aigner, A. R. The phenomenon of strain degeneration in biotechnologically relevant fungi. Appl Microbiol Biotechnol 107, 4745–4758 (2023).
Hyde, K. D. et al. Current trends, limitations and future research in the fungi? Fungal Diversity 125, 1–71 (2024).
Pye, C. R., Bertin, M. J., Lokey, R. S., Gerwick, W. H. & Linington, R. G. Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl. Acad. Sci. U.S.A. 114, 5601–5606 (2017).
Ahrendt, S. R., Mondo, S. J., Haridas, S. & Grigoriev, I. V. MycoCosm, the JGI’s Fungal Genome Portal for Comparative Genomic and Multiomics Data Analyses. in Microbial Environmental Genomics (MEG) (eds. Martin, F. & Uroz, S.) vol. 2605 271–291 (Springer US, New York, NY, 2023).
Grigoriev, I. V. et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucl. Acids Res. 42, D699–D704 (2014).
Crous, P. W., Grams, W., Stalpers, J. A., Robert, V. & Stegehuis, G. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50, 19–22 (2004).
Robert, V. et al. MycoBank gearing up for new horizons. IMA Fungus 4, 371–379 (2013).
Huberman, L. B. Developing Functional Genomics Platforms for Fungi. mSystems 6, https://doi.org/10.1128/msystems.00730-21 (2021).
Hyde, K. D. The numbers of fungi. Fungal Diversity 114, 1–1 (2022).
Paterson, R. R. M., Solaiman, Z. & Santamaria, O. Guest edited collection: fungal evolution and diversity. Sci Rep 13, 21438, s41598-023-48471–0 (2023).
Wu, B. et al. Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology 10, 127–140 (2019).
Aime, M. C. & Brearley, F. Q. Tropical fungal diversity: closing the gap between species estimates and species discovery. Biodivers Conserv 21, 2177–2180 (2012).
Stallman, J. K. et al. The contribution of tropical long-term studies to mycology. IMA Fungus 15, 35 (2024).
Chisholm, R. A. et al. Two centuries of biodiversity discovery and loss in Singapore. Proc. Natl. Acad. Sci. USA 120, e2309034120 (2023).
Davison, G. Special Ecology Feature: Biodiversity’s Crucial Role in the Modern Singapore City. CITYGREEN 01, 102 (2012).
Choong, M. F. A. Education on plants and fungi in Singapore: an urgent call. Nature in Singapore Supplement 1, 279286 (2022).
Lee, S. & Choong, A. Checklist of Fungi Species with their Category of Threat Status for Singapore. in The Singapore red data book: red lists of Singapore biodiversity 515–518 (National Parks Board, Singapore, 2024).
Weerakoon, G., Ngo, K. M., Lum, S., Lumbsch, H. T. & Lücking, R. On time or fashionably late for lichen discoveries in Singapore? Seven new species and nineteen new records of Graphidaceae from the Bukit Timah Nature Reserve, a highly urbanized tropical environment in South-East Asia. The Lichenologist 47, 157–166 (2015).
Katoh, K. & Standley, D. M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30, 772–780 (2013).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 5, e9490 (2010).
Yu, G., Smith, D. K., Zhu, H., Guan, Y. & Lam, T. T. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8, 28–36 (2017).
He, K., Zhang, X., Ren, S. & Sun, J. Deep Residual Learning for Image Recognition. in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778, https://doi.org/10.1109/CVPR.2016.90 (IEEE, Las Vegas, NV, USA, 2016).
Zhou, X. et al. EAST: An Efficient and Accurate Scene Text Detector. in 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2642–2651, https://doi.org/10.1109/CVPR.2017.283 (IEEE, Honolulu, HI, 2017).
Eliahu, N., Igbaria, A., Rose, M. S., Horwitz, B. A. & Lev, S. Melanin Biosynthesis in the Maize Pathogen Cochliobolus heterostrophus Depends on Two Mitogen-Activated Protein Kinases, Chk1 and Mps1, and the Transcription Factor Cmr1. Eukaryot Cell 6, 421–429 (2007).
Plemenitaš, A., Vaupotič, T., Lenassi, M., Kogej, T. & Gunde-Cimerman, N. Adaptation of extremely halotolerant black yeast Hortaea werneckii to increased osmolarity: a molecular perspective at a glance. Studies in Mycology 61, 67–75 (2008).
Ng, S. B. et al. The 160K Natural Organism Library, a unique resource for natural products research. Nat Biotechnol 36, 570–573 (2018).
Seemann, T. barrnap 0.9: rapid ribosomal RNA prediction. https://github.com/tseemann/barrnap (2013).
Paradis, E. & Schliep, K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019).
Ten, D. W. X., Wong, F. T., Lim, Y. H. & Koh, L. C. W. A Singapore-centric Fungal Dataset of 518 Cultivated Strains with Visual Phenotypes and Taxonomic Identity. figshare https://doi.org/10.6084/m9.figshare.29434469 (2025).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP647581 (2025).
Acknowledgements
This work is supported by the Agency for Science, Technology and Research (A*STAR) under C233017006, and the Singapore Integrative Biosystems and Engineering Research Strategic Research & Translational Thrust (SIBER SRTT, A*STAR). The authors extend their gratitude towards Dr. Siew Bee Ng (SIFBI, A*STAR) for her support on strains retrieval.
Author information
Authors and Affiliations
Contributions
F.T.W., Y.H.L. and W.K. conceptualised, designed and coordinated the study. D.W.X.T conducted the experiments and data acquisition. D.W.X.T. wrote the manuscript along with inputs from all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ten, D.W.X., Wong, F.T., Lim, Y.H. et al. A Singapore-centric Fungal Dataset of 518 Cultivated Strains with Visual Phenotypes and Taxonomic Identity. Sci Data (2026). https://doi.org/10.1038/s41597-025-06532-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-06532-1