Abstract
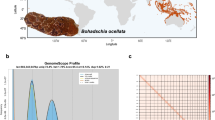
By the integration of MGI short-read sequencing, PacBio HiFi long-read sequencing, and Hi-C techniques, we constructed a chromosome-scale genome assembly for Holothuria fuscocinerea, a highly abundant and widely distributed sea cucumber species in the family Holothuriidae. The assembly was anchored into 23 pseudochromosomes with a total size of 1.556 Gb after redundancy removal, demonstrating contig and scaffold N50 values of 5.18 Mb and 63.82 Mb, respectively, indicative of high contiguity. Genome quality assessment using Merqury and BUSCO analyses yielded a QV score of 60.4087 and a BUSCO completeness rate of 97.8%, confirming high assembly quality with minimal gaps. Among the 29,878 protein coding genes, 95 percent obtained functional annotations from at least one database. This high-resolution genome resource is expected to advance research on H. fuscocinerea and the Holothuroidea class, providing insights for population management, evolutionary studies, and genetics research.
Similar content being viewed by others
Data availability
The following sites store all the data generated in this study available for downloading. The BioProject PRJNA125024973 of NCBI has been created to store raw data acquired in various sequencings and the final H. fuscocinerea genome assembled into 23 chromosomes. Additionally, the accession number JBNFMV00000000074 of DDBJ/ENA/GenBank store the genome assembly. The Figshare database75 hosts all data generated from genome assembly, gene annotation and functional annotations at the link: https://figshare.com/s/6c5acac389a19d8c8386.
Code availability
The following packages, under their established procedures and guidelines, were applied to compute and analyze the data in this study. The workflows and scripts were not altered unless otherwise specified. No custom code was used in this study for data processing, curation, or validation. All analyses were performed using publicly available tools as described in the Methods section. The detailed list of parameters of each software is as follows:
SOAPnuke (v2.1.4): Applied to remove low-quality reads from MGI sequencing raw reads using built-in configurations.
SMRT Link (v13.1): Applied to transform and filter PacBio sequencing raw data using built-in configurations.
Jellyfish (v2.3.0): Applied to count K-mers for assessment of genome size and heterozygosity with a K-mer size of 21.
GenomeScope (v2.0.0): Applied to process the K-mer histogram for the assessment of genome size, heterozygosity and repetitive component using built-in configurations.
Hifiasm (v0.19.8-r603): Applied to assemble the PacBio HiFi data after reads comparison and self-correct using built-in configurations.
Bwa (v0.7.17-r1188): Applied to map the MGI short read data onto the draft assembly using built-in configurations.
Pilon (v1.23): Applied to correct residual errors with Bwa alignment result using built-in configurations.
Purge_dups (v1.2.5): Applied to reduce redundant haplotigs and determine heterozygosity for the draft genome under a configuration of -j 80 -s 80.
ALLHiC (v1.1): Applied to assign and orient scaffolds using Hi-C reads into chromosome-level assemblies.
Merqury (v1.3): Applied to assess k-mer coverage and QV value for the qualification of the assembled genome using best-fit K-mer = 19.
BUSCO (v5.7.1): Applied to estimate genomic coverage using the metazoa_odb10 data collection.
Circos (v0.69): Applied to display chromosomal structure and visualize the distribution of gene regions, repeat sequences, SNP percentage and NGS sequencing depth.
RepeatMasker (v4.09): Applied to annotate transposable elements using built-in configurations.
EDTA: Applied to annotate de-novo transposable elements using built-in configurations.
Barrnap (v0.9): Applied to identify ribosomal RNAs (rRNAs) using built-in configurations.
tRNAscan-SE (v2.0.11): Applied to search for transfer RNAs (tRNAs) sing built-in configurations.
Infernal (v1.1.4): Applied to identify microRNAs (miRNAs) and small nuclear RNAs (snRNAs) using built-in configurations.
Braker (v3.0.8): Applied to integrate gene prediction results with 9 selected proteomes and RNAseq reads from tissues with parameters set to gff3, threads 48, prot_seq = pep.fasta, bam = bams and UTR = on.
HISAT2 (v2.2.1): Applied to map transcriptomic data for genome annotation using built-in configurations.
StringTie (v2.1.7): Applied to assemble the transcripts for the prediction of gene structures using built-in configurations.
MAKER3 (v3.01.03): Applied to combine outputs from various prediction modes into the final gene collection using built-in configurations.
BLAST (v2.11.0+): Applied for synteny analysis and functional annotation of predicted genes with blastp module set to e-value = 1e^-5.
References
Conand, C. Expansion of global sea cucumber fisheries buoys exports. Revista de Biología Tropical 65, S1–S10–S11–S10 (2017).
Miller, A. K. et al. Molecular phylogeny of extant Holothuroidea (Echinodermata). Molecular Phylogenetics and Evolution 111, 110–131 (2017).
Conand, C. Tropical sea cucumber fisheries: Changes during the last decade. Mar Pollut Bull 133, 590–594 (2018).
Purcell S., Conand C., Uthicke S., Byrne M. Ecological Roles of Exploited Sea Cucumbers. In: Oceanography and Marine Biology An Annual Review V54) (2016).
Hamamoto, K., Poliseno, A., Soliman, T. & Reimer, J. D. Shallow epifaunal sea cucumber densities and their relationship with the benthic community in the Okinawa Islands. PeerJ 10, e14181 (2022).
Zixuan, E. et al. Applications of Environmental DNA (eDNA) in Monitoring the Endangered Status and Evaluating the Stock Enhancement Effect of Tropical Sea Cucumber Holothuria Scabra. Marine biotechnology (New York, NY) 25, 778–789 (2023).
Huang, W. et al. Spawning, larval development and juvenile growth of the tropical sea cucumber Holothuria leucospilota. Aquaculture 488, 22–29 (2018).
Purcell, S. W. et al. Commercially important sea cucumbers of the world. Food & Agriculture Org., (2023).
Conand C., Gamboa, R. & Purcell, S. Holothuria fuscocinerea. The IUCN Red List of Threatened Species 2013: e.T180494A1638255.) (2013).
G P. Holothuria (Stauropora) fuscocinerea Jaeger, 1833..). World Register of Marine Species (2018).
Dermawan A. Pedoman umun identifikasi dan monitoring Teripang. Direktorat Konservasi dan Keanekaragaman Hayati Laut Jakarta, 8-56 (2015).
Demeuldre, M. et al. Mechanical adaptability of sea cucumber Cuvierian tubules involves a mutable collagenous tissue. J Exp Biol 220, 2108–2119 (2017).
Cheng, C. et al. Characterization and phylogenetic analysis of the complete mitochondrial genome of a tropical sea cucumber, Holothuria fuscocinerea. Mitochondrial DNA Part B 5, 2677–2678 (2020).
Sun, Y. et al. The complete mitochondrial genome of Holothuria fuscocinerea (Jaeger,1833). Mitochondrial DNA B Resour 5, 33–34 (2019).
Anderson, S. C., Flemming, J. M., Watson, R. & Lotze, H. K. Serial exploitation of global sea cucumber fisheries. Fish and Fisheries 12, 317–339 (2011).
Zhang, S.-Y., Yi, Y.-H. & Tang, H.-F. Bioactive triterpene glycosides from the sea cucumber Holothuria fuscocinerea. Journal of natural products 69, 1492–1495 (2006).
Cayabo, G. D. B. & Mabuhay-Omar, J. A. Antibacterial potential of crude extracts from sea cucumber Holothuria fuscoscinerea Jaeger, 1833. The Palawan Scientist 8, 1–1 (2016).
Siregar, M. S. A., Bachtiar, E., Nurhayati, A. & Lewaru, M. W. Antioxidant Activity of Gamat (Stichopus variegatus) and Milk Sea Cucumbers (Holothuria fuscocinerea) from the Thousand Islands National Park Waters. Journal of Aquaculture and Fish Health 12, 390–404 (2023).
Komala R., Miarsyah M. & Wulaningsih R. Holothuroidea as a constituent of benthic communities in the seagrass ecosystems at Bira Island Islands. In: IOP Conference Series: Earth and Environmental Science). IOP Publishing (2019).
Arriesgado, E. et al. Diversity and abundance of sea cucumbers in selected areas of Mindanao, Philippines. Philippine Journal of Science 151, 863–877 (2022).
Patantis, G., Dewi, A. S., Fawzya, Y. N. & Nursid, M. Identification of Beche-de-mers from Indonesia by molecular approach. Biodiversitas Journal of Biological Diversity 20, 537–543 (2019).
Bilan, M. I. et al. The Structure of Sulfated Polysaccharides from the Sea Cucumber Holothuria (Stauropora) fuscocinerea. Russian Journal of Bioorganic Chemistry 49, 758–767 (2023).
Morton, B. Aspects of predation by Tonna zonatum (Prosobranchia: Tonnoidea) feeding on holothurians in Hong Kong. Journal of molluscan studies 57, 11–19 (1991).
López-Pérez, A., Pérez-López, J., Granja-Fernández, R., Calva-Benítez, L.-G. & Maya-Alvarado, B. Feeding habits of Holothuria (Stauropora) fuscocinerea (Echinodermata: Holothuroidea) in a Mexican Pacific reef. Revista de Biología Tropical 69, 66–79 (2021).
Bondoc, K. G. V., Lee, H., Cruz, L. J., Lebrilla, C. B. & Juinio-Meñez, M. A. Chemical fingerprinting and phylogenetic mapping of saponin congeners from three tropical holothurian sea cucumbers. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 166, 182–193 (2013).
Pangestuti R. et al. Free Radical Scavenging Activity of Selected Sea Cucumber Species from Mataram-Lombok, Indonesia. Jurnal Teknologi 78 (2016).
Sun, L., Jiang, C., Su, F., Cui, W. & Yang, H. Chromosome-level genome assembly of the sea cucumber Apostichopus japonicus. Scientific Data 10, 454 (2023).
Chen, T. et al. The Holothuria leucospilota genome elucidates sacrificial organ expulsion and bioadhesive trap enriched with amyloid-patterned proteins. Proceedings of the National Academy of Sciences 120, e2213512120 (2023).
Zhong, S. et al. Chromosomal-level genome assembly and annotation of the tropical sea cucumber Holothuria scabra. Scientific Data 11, 474 (2024).
Chen, T. et al. Chromosome-level genome assembly and annotation of the tropical sea cucumber Stichopus monotuberculatus. Scientific Data 11, 1245 (2024).
Medina-Feliciano, J. G., Pirro, S., García-Arrarás, J. E., Mashanov, V. & Ryan, J. F. Draft genome of the sea cucumber Holothuria glaberrima, a model for the study of regeneration. Frontiers in Marine Science 8, 603410 (2021).
Borrero-Perez, G. H., Gomez-Zurita, J., Gonzalez-Wanguemert, M., Marcos, C. & Perez-Ruzafa, A. Molecular systematics of the genus Holothuria in the Mediterranean and Northeastern Atlantic and a molecular clock for the diversification of the Holothuriidae (Echinodermata: Holothuroidea). Mol Phylogenet Evol 57, 899–906 (2010).
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010) (2017).
Chen, Y. et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7, gix120 (2018).
Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nature biotechnology 37, 1155–1162 (2019).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Vurture, G. W. et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204 (2017).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature methods 18, 170–175 (2021).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. bioinformatics 25, 1754–1760 (2009).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS one 9, e112963 (2014).
Guan, D. et al. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 36, 2896–2898 (2020).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome research 19, 1639–1645 (2009).
Uliano-Silva, M. et al. MitoHiFi: a python pipeline for mitochondrial genome assembly from PacBio high fidelity reads. BMC bioinformatics 24, 288 (2023).
Ou, S. et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome biology 20, 1–18 (2019).
Ellinghaus, D., Kurtz, S. & Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC bioinformatics 9, 18 (2008).
Ou, S. & Jiang, N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant physiology 176, 1410–1422 (2018).
Su, W., Gu, X. & Peterson, T. TIR-Learner, a new ensemble method for TIR transposable element annotation, provides evidence for abundant new transposable elements in the maize genome. Molecular plant 12, 447–460 (2019).
Xiong, W., He, L., Lai, J., Dooner, H. K. & Du, C. HelitronScanner uncovers a large overlooked cache of Helitron transposons in many plant genomes. Proceedings of the National Academy of Sciences 111, 10263–10268 (2014).
Shi, J. & Liang, C. Generic repeat finder: a high-sensitivity tool for genome-wide de novo repeat detection. Plant physiology 180, 1803–1815 (2019).
Smit A. F. A. HRGP. RepeatModeler Open-1.0. 2008–2015. (ed Biology IfS) (2018).
Aylward, F. Introduction to Prokaryotic gene prediction (CDS and rRNA) V. 2. BMC Bioinformatics 11, 1 (2010).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids research 25, 955–964 (1997).
Kalvari, I. et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic acids research 46, D335–D342 (2018).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013).
Cantarel, B. L. et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome research 18, 188–196 (2008).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology 37, 907–915 (2019).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature biotechnology 33, 290–295 (2015).
Warner, J. F. et al. Chromosomal-level genome assembly of the painted sea urchin Lytechinus pictus: a genetically enabled model system for cell biology and embryonic development. Genome Biology and Evolution 13, evab061 (2021).
Telmer, C. A. et al. Echinobase: a resource to support the echinoderm research community. Genetics 227, iyae002 (2024).
Hall, M. R. et al. The crown-of-thorns starfish genome as a guide for biocontrol of this coral reef pest. Nature 544, 231–234 (2017).
Davidson, P. L. et al. Chromosomal-level genome assembly of the sea urchin Lytechinus variegatus substantially improves functional genomic analyses. Genome Biology and Evolution 12, 1080–1086 (2020).
Sodergren, E. et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science 314, 941–952 (2006).
Brůna, T., Hoff, K. J., Lomsadze, A., Stanke, M. & Borodovsky, M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR genomics and bioinformatics 3, lqaa108 (2021).
Nachtweide S., Stanke M. Multi-genome annotation with AUGUSTUS. In: Gene prediction: methods and protocols). Springer (2019).
Stover, N. A. & Cavalcanti, A. R. Using NCBI BLAST. Current Protocols Essential Laboratory Techniques 14, 11.11. 11–11.11. 34 (2017).
Boutet E. et al. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Plant bioinformatics: methods and protocols, 23-54 (2016).
UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic acids research 46, 2699–2699 (2018).
Kanehisa, M., Sato, Y. & Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. Journal of molecular biology 428, 726–731 (2016).
Mitchell, A. L. et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic acids research 47, D351–D360 (2019).
Consortium GO. The gene ontology resource: 20 years and still GOing strong. Nucleic acids research 47, D330–D338 (2019).
Brown, M. R., Manuel Gonzalez de La Rosa, P. & Blaxter, M. Tidk: a toolkit to rapidly identify telomeric repeats from genomic datasets. Bioinformatics 41, btaf049 (2025).
Lin, Y. et al. quarTeT: a telomere-to-telomere toolkit for gap-free genome assembly and centromeric repeat identification. Horticulture research 10, uhad127 (2023).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP579817 (2025).
T C. Holothuria fuscocinerea isolate:south_sea_zhhs | cultivar:nanhai Genome sequencing and assembly). GenBank. https://identifiers.org/ncbi/insdc:JBNFMV000000000 (2025).
T C. Chromosome-level genome assembly and annotation of the tropical sea cucumber Holothuria fuscocinerea). Figshare. https://doi.org/10.6084/m9.figshare.28831169 (2025).
Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome biology 21, 1–27 (2020).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell systems 3, 99–101 (2016).
Acknowledgements
This study was graciously supported by grants from the National Natural Science Foundation of China (42176132 and 32573487 to T.C., and W2512089 to L.S. and A.Y.), the Research on breeding technology of candidate species for Guangdong modern marine ranching (2024-MRB-00-001 to T.C.), the Science and Technology Program of Nansha District (NSJL202103 to C.H.),, the National Key R & D Program of China (2022YFD2401301 to C.H.), the Guangdong Province Project (2024A1515011418 to T.C., 2024A1515010899 to X.J.).
Author information
Authors and Affiliations
Contributions
Chunhua Ren, Chaoqun Hu, Aifen Yan, and Ting Chen planned and conceptualized the research. Xuan Wang, Qianying Huang, Zhou Qin, Wenjie Pan, Jiasheng Huang, Zepeng Zhang, Hua Ge, Jingxuan Liang, Jianfeng Xu, and Yi Zhang acquired and processed the samples. Zhou Qin and Dingding Fan constructed the genome and performed annotations. Xuan Wang, Qianying Huang, Zhou Qin and Dingding Fan analysed gene functions. Xuan Wang, Qianying Huang, Zhou Qin, Dingding Fan and Ting Chen conducted bioinformatic analyses. Chunhua Ren, Peng Luo, Xiao Jiang, Lina Sun, Hongyan Sun, Chaoqun Hu, Aifen Yan, and Ting Chen offered experimental materials and computational resources. Xuan Wang, Qianying Huang, Dingding Fan, Aifen Yan and Ting Chen composed the manuscript. Chunhua Ren, Chaoqun Hu, Aifen Yan, and Ting Chen carried out revisions. All authors have reviewed and consented to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, X., Huang, Q., Qin, Z. et al. Chromosome-level genome assembly and annotation of the tropical sea cucumber Holothuria fuscocinerea. Sci Data (2026). https://doi.org/10.1038/s41597-026-06609-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-026-06609-5