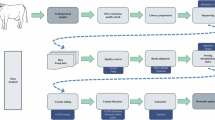
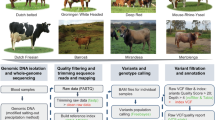
Abstract
China’s abundant indigenous yellow cattle resources are of great significance for studying environmental adaptability evolution, genetic resource conservation, and breeding improvement. The majority of the cattle population consists of indigenous breeds. Understanding the genetic architecture of these cattle breeds is essential for effective management and conservation efforts. In this study, we collected DNA samples from five local cattle breeds (n = 56) and obtained whole-genome sequencing (WGS) data for 10 Jinchuan (JC) yak samples from the NCBI database as the outgroup. Whole-genome resequencing generated approximately 2.3 TB of paired-end data, achieving an average depth of 13X and a depth range of 9.75X to 39.03X across the 66 samples. The sequencing data were pre-processed and mapped to the cattle reference genome (ARS-UCD1.2) with an alignment rate of 99.5%. Finally, the variant calling process produced approximately 31 million high-quality SNPs. These data enhance our understanding of cattle genetic architecture, enabling the discovery of functional variants and evolutionary insights to inform breeding strategies for climate-resilient and sustainable cattle production.
Similar content being viewed by others
Data availability
The raw sequencing data generated in this study have been deposited in both the NCBI Sequence Read Archive under BioProject accession PRJNA1369724 (SRA: SRP655464)21 and the China National GeneBank DataBase (CNGBdb) under accession number CNP000755222. The final variant sets (including SNPs, InDels, SVs, and CNVs) are available in the Figshare repository23.
Code availability
Data analyses were primarily performed using standard bioinformatics tools within a Linux operating system environment. Detailed information regarding software versions and parameter settings is available at: https://github.com/triple-y/WGS-Chinese-yellow-cattle.
References
Lyu, Y. et al. Recent selection and introgression facilitated high-altitude adaptatio n in cattle. Sci Bull (Beijing) 69, 3415–3424, https://doi.org/10.1016/j.scib.2024.05.030.
Zhang, G. X. et al. Genetic diversity and population structure of indigenous yellow cattle breeds of China using 30 microsatellite markers. Anim Genet 38, 550–559, https://doi.org/10.1111/j.1365-2052.2007.01644.x.
Friedrich, J. & Wiener, P. Selection signatures for high-altitude adaptation in ruminants. Anim Genet 51, 157-165, https://doi.org/10.1111/age.12900.
Gong, Y., Li, Y., Liu, X., Ma, Y. & Jiang, L. A review of the pangenome: how it affects our understanding of genomic variation, selection and breeding in domestic animals? J Anim Sci Biotechnol 14, 73, https://doi.org/10.1186/s40104-023-00860-1.
Zhang, Y.-W., Wu, S.-X., Wang, G.-W., Wan, R.-D. & Yang, Q.-E. Single-cell analysis identifies critical regulators of spermatogonial development and differentiation in cattle-yak bulls. J Dairy Sci 107, 7317–7336, https://doi.org/10.3168/jds.2023-24442.
Liu, Y. et al. Discovery of Genomic Characteristics and Selection Signatures in Southern Chinese Local Cattle. Front Genet 11, 533052, https://doi.org/10.3389/fgene.2020.533052.
Zhang, W. et al. Genome-wide assessment of genetic diversity and population structure i nsights into admixture and introgression in Chinese indigenous cattle. BMC Genet 19, 114, https://doi.org/10.1186/s12863-018-0705-9.
Moore, A. J. & Kukuk, P. F. Quantitative genetic analysis of natural populations. Nat Rev Genet 3, 971–978, https://doi.org/10.1038/nrg951.
Li, Z. et al. Impacts of SNP genotyping call rate and SNP genotyping error rate on i mputation accuracy inHolsteincattle. Yi Chuan 41, 644-652, https://doi.org/10.16288/j.yczz.18-319.
Hulsegge, I., Oldenbroek, K., Bouwman, A., Veerkamp, R. & Windig, J. Selection and Drift: A Comparison between Historic and Recent Dutch Fr iesian Cattle and Recent Holstein Friesian Using WGS Data. Animals (Basel) 12, 329, https://doi.org/10.3390/ani12030329.
Xia, X. et al. Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data. BMC Genomics 22, 43, https://doi.org/10.1186/s12864-020-07340-0.
Yang, B. et al. Genetic Diversity Estimation and Genome-Wide Selective Sweep Analysis of the Bazhou Yak. Animals (Basel) 15, 849, https://doi.org/10.3390/ani15060849.
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884-i890, https://doi.org/10.1093/bioinformatics/bty560.
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 25, 1754-1760, https://doi.org/10.1093/bioinformatics/btp324.
Pham, M., Tu, Y. & Lv, X. Accelerating BWA-MEM Read Mapping on GPUs. ICS 2023, 155-166, https://doi.org/10.1145/3577193.3593703.
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078-2079, https://doi.org/10.1093/bioinformatics/btp352.
Grant, J. R. et al. A large structural variant collection in Holstein cattle and associate d database for variant discovery, characterization, and application. BMC Genomics 25, 903, https://doi.org/10.1186/s12864-024-10812-2.
Abyzov, A., Urban, A. E., Snyder, M. & Gerstein, M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21, 974–984, https://doi.org/10.1101/gr.114876.110.
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next- generation DNA sequencing data. Genome Res 20, 1297-1303, https://doi.org/10.1101/gr.107524.110.
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughpu t sequencing data. Nucleic acids research 38, e164, https://doi.org/10.1093/nar/gkq603.
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP655464 (2025).
Wenjie, Han. Population Genetic Analysis of Chinese indigenous cattle. China National GeneBank Database https://doi.org/10.26036/CNP0007552 (2025).
Wang, W. Population Genetic Analysis of Chinese indigenous cattle. figshare https://doi.org/10.6084/m9.figshare.30759347 (2025).
Acknowledgements
This work was supported by the Sichuan Province Science and Technology Planning Project (2021YFYZ0001), the “5 + 1” Special Project for Breakthroughs in Cutting-Edge Agricultural Technologies (5 + 1QYGG003), the Sichuan Beef Cattle Innovation Team Project (SCCXTD-2025-13), the Sichuan Fiscal Operations Special Program (SASA2025CZYX003), the Basic Research Projects of Scientific Research Institutes (SASA202505), and the Sichuan Province Science and Technology Support Project (2024ZYD0283).
Author information
Authors and Affiliations
Contributions
W.W. and J.Y. conceived and designed the study. W.W. were involved in the review and writing process, L.L., Y.C., X.M., Y.A., J.G., D.F., X.D., X.C., F.H., Y.S., C.W., Z.Y., M.F. performed experiments and analyzed data. W.W. and J.Y. supervised the project and acquired funding. All authors made critical contributions to the manuscript drafts.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, W., Li, L., Chen, Y. et al. Whole-genome resequencing and genetic diversity of five indigenous cattle breeds from China. Sci Data (2026). https://doi.org/10.1038/s41597-026-06610-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-026-06610-y