Abstract
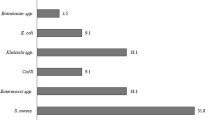
Bloodstream infections (BSIs) of high morbidity and mortality are across all age groups, and urgent for accurate intervention. Gram stain interpretation of positive blood cultures (PBCs) is crucial for early diagnosing BSIs, yet this manual process is labor-intensive, time-consuming, and highly operator-dependent. Artificial intelligence (AI)-assisted microscopic interpretation of stained smears presents beneficial to microbiology diagnostics. Addressing the auto-identification of blood-culture Gram stains, this study introduces a dataset of Gram-stain smears collected in clinical practice. The dataset includes 505 microscopic images, covering up to 57 species associated with BSIs, with a total of 7528 annotations. These annotations categorized by staining characteristics and morphological features into cocci, bacilli, and fungi. We trained and validated an object detection model based on the YOLOv10 architecture on this dataset to automatically localize and classify these morphological categories in microscopic images. The publicly released dataset will help developments that utilize artificial intelligence to auto-interpretate the Gram stains from PBCs for routine clinical application.
Similar content being viewed by others
Data availability
The dataset is available at the Figshare repository16.
Code availability
The annotation tool used for the dataset labelling is publicly available in GitHub, https://github.com/jsbroks/coco-annotator/. The customizable Image Annotation Tools used for the dataset labelling technical check is available from https://github.com/KeyOfSpectator/ImageAnnotationTools, including Double Check IoU Annotation Tool and COCO Json Merge/Split Tool.
References
Jin, L. et al. Clinical Profile, Prognostic Factors, and Outcome Prediction in Hospitalized Patients With Bloodstream Infection: Results From a 10-Year Prospective Multicenter Study. Front Med (Lausanne) 8 (2021).
Dubourg, G., Raoult, D. & Fenollar, F. Emerging methodologies for pathogen identification in bloodstream infections: an update. Expert Rev Mol Diagn 19, 161–173 (2019).
Adrie, C. et al. Attributable mortality of ICU-acquired bloodstream infections: Impact of the source, causative micro-organism, resistance profile and antimicrobial therapy. J Infect 74, 131–141 (2017).
Ikuta, K. S. et al. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. The Lancet 400, 2221–2248 (2022).
Timsit, J. F., Ruppé, E., Barbier, F., Tabah, A. & Bassetti, M. Bloodstream infections in critically ill patients: an expert statement. Intensive Care Med 46, 266–284 (2020).
Cecconi, M., Evans, L., Levy, M. & Rhodes, A. Sepsis and septic shock. The Lancet 392, 75–87 (2018).
Kern, W. V. & Rieg, S. Burden of bacterial bloodstream infection—a brief update on epidemiology and significance of multidrug-resistant pathogens. Clinical Microbiology and Infection 26, 151–157 (2020).
Pien, B. C. et al. The clinical and prognostic importance of positive blood cultures in adults. American Journal of Medicine 123, 819–828 (2010).
Lamy, B., Sundqvist, M. & Idelevich, E. A. Bloodstream infections – Standard and progress in pathogen diagnostics. Clinical Microbiology and Infection 26, 142–150 (2020).
Ito, H. et al. The role of Gram stain in reducing broad-spectrum antibiotic use: A systematic literature review and meta-analysis. Infect Dis Now 53, 104764 (2023).
Thomson, R. B. One small step for the Gram stain, one giant leap for clinical microbiology. J Clin Microbiol 54, 1416–1417 (2016).
Smith, K. P. & Kirby, J. E. Image analysis and artificial intelligence in infectious disease diagnostics. Clinical Microbiology and Infection 26, 1318–1323 (2020).
Smith, K. P., Kang, A. D. & Kirby, J. E. Automated interpretation of blood culture Gram stains by use of a deep convolutional neural network. J Clin Microbiol 56 (2018).
Walter, C. et al. Performance evaluation of machine-assisted interpretation of Gram stains from positive blood cultures. J Clin Microbiol 62 (2024).
Makrai, L. et al. Annotated dataset for deep-learning-based bacterial colony detection. Sci Data 10, 497 (2023).
Yi, Q. et al. An annotated dataset of Gram stains from positive blood cultures. Figshare. https://doi.org/10.6084/m9.figshare.26004610
Acknowledgements
We appreciate the help from Mr. Shichun Feng for technical validation. This work has been supported by the Noncommunicable Chronic Diseases-National Science and Technology Major Project [No.2024ZD0532800], Peking Union Medical College Hospital Talent Cultivation Program-Category D [No. UHB12289], and Young Elite Scientists Sponsorship Program of the Beijing High Innovation Plan.
Author information
Authors and Affiliations
Contributions
Q.Y. and Y.X. conceived the concept of the work. X.G., M.H. X.W. and T.W. performed the microorganism identification. Q.Y., X.G., R.Z. K.X. and X.X. made the digital images and annotated the images. R.Z. K.X. and X.X. curated the digital images. Q.Y. drafted the manuscript and performed the technical validation. Q.Y. and Y.X. had overarching administrative responsibility for the project. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yi, Q., Gou, X., Zhu, R. et al. An annotated dataset of Gram stains from positive blood cultures. Sci Data (2026). https://doi.org/10.1038/s41597-026-06651-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-026-06651-3