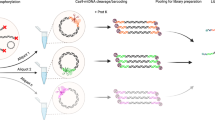
Abstract
Mitochondrial DNA (mtDNA) mutations are critical to disease research, evolutionary studies, and lineage tracing but are challenging to analyze due to interference from nuclear mitochondrial sequences (NUMTs). Current high-throughput sequencing techniques rely on multiple primers or probes to amplify short mtDNA fragments, followed by alignment to a reference genome. However, this approach fails to mitigate NUMTs interference, leading to ambiguous results. In this study, we presented a nanopore-based third-generation sequencing (TGS) method using a single primer pair to amplify full-length mtDNA, effectively circumventing NUMTs artifacts. Sequencing was carried out on the QITAN TECH QNome-3841hex platform, generating complete mtDNA coverage for 106 samples from eight distinct family pedigrees, including complex familial structures such as half-siblings and multi-generational households. The sequencing achieved 100% genome coverage with an average mapping rate of 99.96%, supporting comprehensive genome characterization. The resulting dataset offers valuable insights into mtDNA mutation detection, mitochondrial genetics, population genetics, ancestry tracing, and forensic identification, and may advance mtDNA sequencing technologies and intergenerational studies.
Similar content being viewed by others
Data availability
All raw sequencing data generated in this study are available in the NCBI Sequence Read Archive (SRA) under accession SRP57037539. Consensus mitochondrial genome sequences are available in GenBank via BioProject PRJNA123594740. Variant data are available in the European Variation Archive (EVA) under PRJNA123594741. Additional supporting data, including sample metadata, pedigree charts, and quality control metrics, are available in Figshare repository24.
Code availability
All analyses were performed using publicly available tools, and no modifications were made to the original software. To ensure reproducibility, all commands and scripts used for FASTQ-based quality control, alignment and variant calling have been consolidated into a reproducible workflow. This workflow is available at https://github.com/myjasminum/mito under an open-source license.
References
Borcherding, N. & Brestoff, J. R. The power and potential of mitochondria transfer. Nature 623, 283–291, https://doi.org/10.1038/s41586-023-06537-z (2023).
Hu, Z. et al. A novel protein CYTB-187AA encoded by the mitochondrial gene CYTB modulates mammalian early development. Cell Metabolism 36, 1586–1597.e7, https://doi.org/10.1016/j.cmet.2024.04.012 (2024).
Ng, Y. S. et al. Mitochondrial disease in adults: Recent advances and future promise. The Lancet Neurology 20, 573–584, https://doi.org/10.1016/S1474-4422(21)00098-3 (2021).
Ng, Y. S. & Turnbull, D. M. Mitochondrial disease: Genetics and management. J Neurol 263, 179–191, https://doi.org/10.1007/s00415-015-7884-3 (2016).
Castellani, C. A. et al. Mitochondrial DNA copy number can influence mortality and cardiovascular disease via methylation of nuclear DNA CpGs. Genome Med 12, 84, https://doi.org/10.1186/s13073-020-00778-7 (2020).
Zhang, H., Zhu, Y. & Xue, D. Moderate embryonic delay of paternal mitochondrial elimination impairs mating and cognition and alters behaviors of adult animals. Sci. Adv. 10, eadp8351, https://doi.org/10.1126/sciadv.adp8351 (2024).
Kristjansson, D., Bohlin, J., Jugessur, A. & Schurr, T. G. Matrilineal diversity and population history of norwegians. American J Phys Anthropol 176, 120–133, https://doi.org/10.1002/ajpa.24345 (2021).
Yue, W. et al. Investigation of control region sequences of mtDNA in naqu tibetan population from northwestern China. Annals of Human Biology 48, 70–77, https://doi.org/10.1080/03014460.2021.1877351 (2021).
James, J. E., Piganeau, G. & Eyre‐Walker, A. The rate of adaptive evolution in animal mitochondria. Molecular Ecology 25, 67–78, https://doi.org/10.1111/mec.13475 (2016).
Faccinetto, C. et al. Internal validation and improvement of mitochondrial genome sequencing using the precision ID mtDNA whole genome panel. Int J Legal Med 135, 2295–2306, https://doi.org/10.1007/s00414-021-02686-w (2021).
Kopinski, P. K., Singh, L. N., Zhang, S., Lott, M. T. & Wallace, D. C. Mitochondrial DNA variation and cancer. Nat Rev Cancer 21, 431–445, https://doi.org/10.1038/s41568-021-00358-w (2021).
Parakatselaki, M.-E. & Ladoukakis, E. D. mtDNA heteroplasmy: Origin, detection, significance, and evolutionary consequences. Life 11, 633, https://doi.org/10.3390/life11070633 (2021).
Zaragoza, M. V., Fass, J., Diegoli, M., Lin, D. & Arbustini, E. Mitochondrial DNA variant discovery and evaluation in human cardiomyopathies through next-generation sequencing. PLoS ONE 5, e12295, https://doi.org/10.1371/journal.pone.0012295 (2010).
Wong, L.-J. C. Diagnostic challenges of mitochondrial DNA disorders. Mitochondrion 7, 45–52, https://doi.org/10.1016/j.mito.2006.11.025 (2007).
Li, M. et al. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. The American Journal of Human Genetics 87, 237–249, https://doi.org/10.1016/j.ajhg.2010.07.014 (2010).
Guo, Y. et al. The effect of strand bias in illumina short-read sequencing data. BMC Genomics 13, 666, https://doi.org/10.1186/1471-2164-13-666 (2012).
Shaw, J., Boucher, C., Yu, Y. W., Noyes, N. & Li, H. Long-read reconstruction of many diverse haplotypes with devider. Genome Res gr. 280510.125, https://doi.org/10.1101/gr.280510.125 (2025).
Macken, W. L. et al. Enhanced mitochondrial genome analysis: Bioinformatic and long-read sequencing advances and their diagnostic implications. Expert Rev Mol Diagn 23, 797–814, https://doi.org/10.1080/14737159.2023.2241365 (2023).
Dayama, G., Emery, S. B., Kidd, J. M. & Mills, R. E. The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res 42, 12640–12649, https://doi.org/10.1093/nar/gku1038 (2014).
Tao, Y., He, C., Lin, D., Gu, Z. & Pu, W. Comprehensive identification of mitochondrial pseudogenes (NUMTs) in the human telomere-to-telomere reference genome. Genes (Basel) 14, 2092, https://doi.org/10.3390/genes14112092 (2023).
Single-cell mitochondrial DNA sequencing: Methodologies and applications. Mitochondrial Communications 2, 107–113, https://doi.org/10.1016/j.mitoco.2024.10.001 (2024).
Wei, W. et al. Nuclear-embedded mitochondrial DNA sequences in 66,083 human genomes. Nature 611, 105–114, https://doi.org/10.1038/s41586-022-05288-7 (2022).
Xue, L., Moreira, J. D., Smith, K. K. & Fetterman, J. L. The mighty NUMT: Mitochondrial DNA flexing its code in the nuclear genome. Biomolecules 13, 753, https://doi.org/10.3390/biom13050753 (2023).
Liu, Y. A full-length mtDNA dataset for studying genetic variations across generations and complex family structures. figshare https://doi.org/10.6084/m9.figshare.30856568 (2025).
Andrews, S. A Quality Control Tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048, https://doi.org/10.1093/bioinformatics/btw354 (2016).
Andrews, R. M. et al. Reanalysis and revision of the cambridge reference sequence for human mitochondrial DNA. Nat Genet 23, 147–147, https://doi.org/10.1038/13779 (1999).
Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100, https://doi.org/10.1093/bioinformatics/bty191 (2018).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008, https://doi.org/10.1093/gigascience/giab008 (2021).
Weissensteiner, H., Forer, L., Kronenberg, F. & Schönherr, S. mtDNA-server 2: Advancing mitochondrial DNA analysis through highly parallelized data processing and interactive analytics. Nucleic Acids Res 52, W102–W107, https://doi.org/10.1093/nar/gkae296 (2024).
Weissensteiner, H. et al. Contamination detection in sequencing studies using the mitochondrial phylogeny. Genome Res. 31, 309–316, https://doi.org/10.1101/gr.256545.119 (2021).
Schönherr, S., Weissensteiner, H., Kronenberg, F. & Forer, L. Haplogrep 3 - an interactive haplogroup classification and analysis platform. Nucleic Acids Res 51, W263–W268, https://doi.org/10.1093/nar/gkad284 (2023).
van Oven, M. & Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 30, E386–394, https://doi.org/10.1002/humu.20921 (2009).
Schneider, V. A. et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27, 849–864, https://doi.org/10.1101/gr.213611.116 (2017).
Smolka, M. et al. Detection of mosaic and population-level structural variants with Sniffles2. Nat Biotechnol 42, 1571–1580, https://doi.org/10.1038/s41587-023-02024-y (2024).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30, 772–780, https://doi.org/10.1093/molbev/mst010 (2013).
Minh, B. Q. et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37, 1530–1534, https://doi.org/10.1093/molbev/msaa015 (2020).
Rambaut, A. FigTree. https://tree.bio.ed.ac.uk/software/figtree/.
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP570375 (2025).
NCBI BioProject https://identifiers.org/ncbi/bioproject:PRJNA1235947 (2025).
EMBL-EBI EVA https://www.ebi.ac.uk/eva/?eva-study=PRJNA1235947 (2025).
Gupta, R. et al. Nuclear genetic control of mtDNA copy number and heteroplasmy in humans. Nature 620, 839–848, https://doi.org/10.1038/s41586-023-06426-5 (2023).
Acknowledgements
This study was supported by National Natural Science Fund of China (82302124), National Key R&D Program of China (2024YFC3306702) and Scientific Research Projects of Science and Technology Commission of Shanghai Municipality (24JG0500500).
Author information
Authors and Affiliations
Contributions
S.Z. and A.C., conception and design. Y.X., J.Z., experiment execution. Y.N. and Q.Y., data analysis. The manuscript was written by Y.N. and Q.Y. and revised by S.Z. and A.C. All authors contributed to review the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Y., Yang, Q., Xuan, Y. et al. A full-length mtDNA dataset for studying genetic variations across generations and complex family structures. Sci Data (2026). https://doi.org/10.1038/s41597-026-06824-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-026-06824-0