Abstract
Both vitamin D deficiency and single nucleotide polymorphisms (SNPs) in the gene encoding the vitamin D receptor (VDR) have been widely reported to associate with susceptibility to polycystic ovarian syndrome (PCOS). A case-control study was conducted to study the influence of vitamin D status and genotpye for 24 SNPs in four genes in the vitamin D pathway (VDR, DBP, CYP27B1, CYP24A1) on PCOS. Statistical analyses were conducted to identify phenotypic and genotypic factors associated with risk of PCOS and to test for interactions between genotype and vitamin D status. PCOS was independently associated with lower age, higher body mass index, lower waist-hip ratio, vitamin D deficiency (serum 25-hydroxyvitamin D concentration <10 ng/mL), lack of outdoor exercise, increased fasting glucose and a family history of PCOS in at least one first degree relative. No statistically significant association was observed between the genotype of any SNP investigated and risk of PCOS, either as a main effect or in interaction with vitamin D status. We report a strong and independent association between vitamin D deficiency and risk of PCOS in Pakistan, that was not modified by genetic variation in the vitamin D pathway.
Similar content being viewed by others
Introduction
Polycystic ovary syndrome (PCOS) is the most common reproductive, endocrine and metabolic disorder in women of reproductive age1. Globally, the prevalence of PCOS ranges from 4 to 21%, as defined using NIH 1990 and Rotterdam 2003 criteria2. Epidemiological data from Pakistan are scarce, but the prevalence of PCOS has been reported to be particularly high among women of South Asian ethnic origin3,4.
PCOS is thought to arise as a result of the interplay between genetic and environmental factors5. A growing body of literature indicates that vitamin D deficiency may be a risk factor: meta-analyses of observational studies reveal independent associations between PCOS and low serum concentrations of 25-hydroxyvitamin D (25[OH]D, the major circulating vitamin D metabolite) as well as single nucleotide polymorphisms [SNPs] in the gene encoding the vitamin D receptor (VDR)6,7,8,9,10,11. However, data relating to vitamin D status among women with PCOS in Pakistan are lacking. Moreover, studies in the field have not yet tested for main effects of SNPs in the genes encoding the 1-alpha hydroxylase enzyme CYP27B1 (which converts 25[OH]D to its active metabolite 1,25-dihydroxy vitamin D) or the vitamin D 24-hydroxylase enzyme CYP24A1 (which converts 25[OH]D to its major inactive catabolite 24R,25-dihydroxy vitamin D). Neither have they tested for interactions between SNPs in the vitamin D pathway and vitamin D status in influencing the risk of PCOS.
We have previously demonstrated that genetic variation in the vitamin D pathway can modify the influence of vitamin D deficiency and supplementation on clinical phenotype in the context of tuberculosis12,13,14. We, therefore, conducted a case control study in Lahore, Pakistan, to determine whether vitamin D deficiency and SNPs in four vitamin D pathway genes might influence the risk of PCOS either as main effects or in interaction with each other.
Results
Participant characteristics
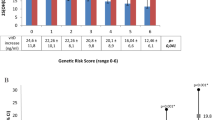
A total of 235 PCOS cases and 235 healthy controls were recruited to the study between April 2016 and April 2018. Characteristics of cases and controls are presented in Table 1. The mean age of cases vs. controls was 25.2 vs 29.7 years, respectively, and the majority (96.8%) of participants were of Punjabi ethnic origin. Married and single women were equally represented in both groups. Students were over-represented, and employed women were under-represented, in cases vs controls, but the proportion of participants at different educational levels did not differ between groups. As anticipated, acne, hair loss, hirsutism, menstrual cycle abnormalities, and pelvic ultrasound scan abnormalities were all more common among cases vs controls. The mean body mass index was higher among cases vs controls, but the mean waist-to-hip ratio was lower. Mean serum concentrations of fasting glucose and luteinizing hormone were higher in cases vs controls, but no difference was seen for mean serum concentration of the follicle-stimulating hormone. As anticipated LH/FSH ratio was higher in cases vs controls. Mean serum 25(OH)D concentration was lower in cases vs controls (17.4 vs 21.7 ng/ml, respectively; P < 0.001).
Association between phenotypic features and risk of PCOS
Table 2 presents the results of univariable and multivariable analyses testing for associations between participant characteristics captured by the study questionnaire or found at physical examination and susceptibility to PCOS. PCOS was independently associated with lower age (adjusted odds ratio [aOR] per additional year of age, 0.86; 95% CI 0.81 to 0.93), higher body mass index (aOR per additional kg/m2, 1.26; 95% CI 1.15 to 1.37), lower waist-hip ratio (aOR per additional unit, 0.88; 95% CI 0.82 to 0.95), vitamin D deficiency (serum 25(OH)D <25 ng/ml, aOR 24.81; 95% CI 6.16 to 99.84), lack of outdoor exercise (aOR 13.47; 95% CI 6.26 to 28.97), increased fasting glucose (aOR per 1 mg/dL increase, 1.12; 95% CI 1.09 to 1.17) and a family history of PCOS in at least one first degree relative (aOR 2.75; 95% CI 1.12 to 6.78). The following features were not found to associate with susceptibility to PCOS: monthly household income, or self-reported consumption of sugary/fast foods, chicken, eggs, milk or red meat.
Association between vitamin D pathway genotype and risk of polycystic ovarian syndrome
Table 3 presents results of univariable and multivariable analyses testing for associations between SNPs in the genes encoding the vitamin D receptor (VDR), the vitamin D 1-alpha hydroxylase enzyme (CYP27B1), the vitamin D 24-hydroxylase enzyme (CYP24A1) and the vitamin D binding protein (DBP) and risk of PCOS. No statistically significant association was observed between the genotype of any polymorphism investigated and risk of PCOS.
Association between vitamin D deficiency and host genotype, stratified by vitamin D status
We next proceeded to investigate whether genetic variation in VDR, CYP24A1, CYP27B1 and DBP modified the association between vitamin D deficiency and risk of PCOS reported above. P values for interaction for these sub-group analyses are presented in Table 3: these revealed no evidence to support the hypothesis that polymorphisms in the vitamin D pathway modify the effect of vitamin D deficiency on risk of PCOS.
Haplotype analysis
We also performed haplotype analysis to assess the cumulative impact of all SNPs on risk of PCOS. The Clark method15 was used to define haplotypes for SNPs showing significant linkage disequilibrium (R2 > 0.80) in the relevant 1000 genomes reference set ([PJL], Punjabi from Lahore, Pakistan). Two SNPs in VDR showed linkage disequilibrium with R2 > 0.80: rs11568820 and rs7976091. Univariate regression analysis for haplotypes TT, TC and CC showed no significant association with the odds of PCOS. Further analysis was performed by adjusting these haplotypes for the phenotypic risk factors for PCOS identified above (age, BMI, WHR, vitamin D status, outdoor physical exercise and fasting glucose). No statistically significant associations with risk of PCOS were identified (Table 4).
Discussion
We report findings from one of the largest and most detailed studies conducted to date to investigate the influence of vitamin D deficiency and genetic variation in the vitamin D pathway on susceptibility to PCOS – and the first such study to be conducted in Pakistan. Uniquely, in addition to investigating SNPs in VDR and DBP, we also explored the influence of variants in CYP24A1 (the gene encoding the major enzymes of 25(OH)D catabolism) and CYP27B1 (the gene encoding the enzyme responsible for converting 25[OH]D to its active metabolite 1,25[OH]2D) on risk of disease. Our major findings were that lower vitamin D status was independently associated with susceptibility to PCOS, but that genetic variation in VDR, DBP, CYP24A1, and CYP27B1 was not. Moreover, we found no evidence of any interaction between serum 25(OH)D levels and genetic variation in the vitamin D pathway on disease risk.
Our finding that vitamin D deficiency associates independently with PCOS risk in women in Pakistan is consistent with reports from other settings including the Netherlands10, Iran16, Egypt17 and the USA18, as well as with findings from a meta-analysis including these and other studies19, showing that mean 25(OH)D level was lower in women with PCOS than controls. The high prevalence of vitamin D deficiency seen among participants in the current study is in keeping with our previous study in Lahore showing a high prevalence of vitamin D deficiency among women of reproductive age20. These and other studies reporting high rates of vitamin D deficiency in other groups21, highlight that vitamin D deficiency is a major public health problem in Pakistan22. The adjusted odds ratio for the association between vitamin D deficiency at the pre-specified 25(OH)D cut-off of 10 ng/ml was higher than has been reported elsewhere; this primarily reflects the very low prevalence of 25(OH)D values <10 ng/ml among controls in the current study. Exploratory analyses investigating 25(OH)D cut-offs of 15 ng/ml (aOR 3.41, 95% CI 1.77 to 6.57), 20 ng/ml (aOR 1.79, 95% CI 0.96 to 3.37), or analyzing 25(OH)D as a continuous variable (aOR 0.98, 95% CI 0.96 to 1.01) showed consistent inverse associations between 25(OH)D level and PCOS risk. Our findings with regard to other factors associated with PCOS, such as higher BMI, reduced physical exercise, higher fasting glucose levels and positive family history are also in keeping with the literature22,23,24,25.
In contrast to these positive findings, the lack of association seen between polymorphisms in VDR in our study contrasts with findings of a recent meta-analysis reporting that the VDR ApaI (rs7975232) polymorphism associated with susceptibility to PCOS in Asian populations (aOR for allelic model, C vs A, 1.19; 95% CI 1.07 to 1.34)6. The lack of association seen for SNPs in DBP is consistent with findings of both the other studies that have investigated polymorphisms in this gene26,27.
Our study has several strengths. Cases comprised a broad range of PCOS phenotypes, including obese and lean, fertile and infertile, and those with and without hyper-androgenic characteristics. We collected detailed information on potential confounders of the association between vitamin D status and PCOS risk, minimizing the potential for residual/unmeasured confounding to explain our findings. We also investigated SNPs in a wider range of vitamin D pathway genes than previously investigated, and applied a stringent correction for multiple testing. By deseasonalizing 25(OH)D data, we were able to calculate individuals average 25(OH)D level throughout the year, which represents their vitamin D status more effectively than a season-specific ‘snapshot’ provided by a single unadjusted reading28.
Our study also has some limitations. Due to the case-control design, reverse causality and/or confounding cannot be excluded as explanations for the associations observed. Although large by comparison with others in the field, our power to detect modest genetic effects, and gene-environment interactions, was limited. 25(OH)D was measured by ELISA rather than with Liquid Chromatography Tandem Mass Spectrometry, which is the gold standard methodology; however, this should not have introduced bias, since the same assay was used to measure vitamin D status in cases and controls. Moreover, the assay we used detects both 25(OH)D2 and 25(OH)D3.
Conclusion
In conclusion, this large case-control study—the first of its kind to be conducted in Pakistan—reports that low vitamin D status associates independently with increased susceptibility to PCOS. However, no statistically significant association between polymorphisms in vitamin D pathway genes and risk of PCOS was demonstrated.
Methods
Study design
We conducted a case control study. Cases were patients aged 14 to 49 years diagnosed with PCOS and recruited from out-patient clinics at the Jinnah Hospital and the Lady Willingdon Hospital, Lahore. A total of 235 women were selected according to Rotterdam criteria, 200329. Patients with thyroid and adrenal diseases and androgen-secreting tumours were excluded. 235 healthy controls were selected from the Citi lab and Research centre, Lahore, on the basis of having no history of infertility, no evidence of clinical hyperandrogenism and normal menstrual cycles. Informed consent was taken from all participants who fulfilled eligibility criteria. Written informed consent was taken from the parents or guardians of the participants who were under 18 years of age. Participants completed a detailed questionnaire including details of age, weight, height, sociodemographic status, dietary habits, physical exercise, sun exposure, androgenic features (acne, hirsutism and patchy hair loss), menstrual cycle history, family history of PCOS and fertility details for married participants. The study was approved by the ethical committee of the University of Punjab (Ref No: Bioethic 125Pu, 13.10.17) and by the ethical review board of Citi lab and Research Centre, Lahore (Ref # 26-17/ERB/CLRC/27th/Dated 28-07-2016). All the methods were performed following the relevant guidelines and regulations approved by the ethical committee.
5 mL of blood was drawn on the 2nd or 3rd day of menstrual cycle30, from a median cubital vein; 2 mL were transferred into vials containing EDTA and frozen at −20 °C for subsequent DNA extraction, and 3 mL was added to serum vials and sent to the laboratory within two hours of collection, where serum was isolated from clotted blood by centrifugation and stored at −20 °C for subsequent determination of 25(OH)D concentration.
Serum 25(OH)D assay
Serum 25(OH) D concentration was determined by ELISA (Calbiotech, EI Cajon U.S.A). Calibrators and controls for the assay were run in duplicates. Interassay CV for serum 25(OH)D for our samples was 14%. Season-adjusted (deseasonalized) values for 25(OH)D were calculated for each participant from their individual standardized 25(OH)D concentration and date of blood sample collection, using a sinusoidal model with values derived from standardized values for all participants as previously described28,31. Vitamin D deficiency was defined using a pre-specified 25(OH)D cut-off of 10 ng/ml; this cut-off was selected a priori on the basis that it is widely used by Public Health bodies32 and that deficiency at this level has been shown to associate most strongly with PCOS11 and other pathologies attributable to vitamin D deficiency33,34,35.
Genotyping
Genomic DNA was extracted from whole blood and quantified using a nanodrop spectrophotometer as previously described36. DNA TaqMan allelic probe assays (Applied Biosystems, Foster City, CA, USA) were used to genotype polymorphisms in genes encoding: the vitamin D receptor (rs4334089, rs10783219, rs4516035, rs11568820 [cdx2], rs7976091, rs731236 [TaqI], rs2228570 [Fok]I, rs1544410 [BsmI], rs7975232 [ApaI], rs7970314, rs2853559, rs2238136); the 1α-hydroxylase enzyme, CYP27B1 (rs4646536, rs4646537); the 24-hydroxylase enzyme, CYP2A41 (rs6013897, rs2762939, rs2248137, rs2762934), and the vitamin D binding protein, DBP (rs7041, rs4588, rs12512631, rs2070741, rs2298849, rs16846876). Taqman® SNP genotyping assays were purchased directly from Life Technologies. Assay IDs are detailed in Supplementary Table 1. One assay was a custom design and primer details are shown in Supplementary Table 2. For SNP Genotyping using the Fluidigm 192.24 Dynamic Array, genomic DNA concentration was normalised to 50 ng/µL and a sample pre-mix was prepared with 2 µL of GTXpress™ Master Mix (2×), 0.2 µL of 20X Fast GT Sample Loading Reagent, 0.2 µL of nuclease-free water and 1.6 µL of genomic DNA. A 10X SNP assay mix was prepared by combining 1.0 µL of Taqman® SNP Genotyping assay with 2.0 µL of 2X Assay Loading Reagent, 0.2uL of ROX™ (50×) and 1.3 µL of nuclease-free water. 3 µL of sample pre-mix and 3 µL of assay mix into each assay inlet of the 192.24 arrays. Pressure fluid was then pipetted into the appropriate wells. The array was then loaded to the Juno™ controller with the RX insert and the Fast PCR 192.24 script run to performs thermal cycling. Thermal conditions were as follows: Hot start 95 °C for 5 minutes, Touchdown (64 °C-61 °C dropping 1 °C per cycle) 4 cycles at 95 °C for 15 seconds, 64 °C 45 for seconds, 72 °C for15 seconds, 34 cycles were run at 95 °C for 15 seconds (denaturing) followed by 60 °C for 45 seconds (annealing) and 72 °C for 15 seconds (extension). Endpoint fluorescent data were collected using the Biomark® Real-Time PCR System, and data were subsequently analysed using the Fluidigm SNP Genotyping Analysis software.
Statistical analysis
Statistical analyses were done with Stata IC (version 15.1). Frequencies of alleles and genotypes were compared using chi-square tests; all were found to be in Hardy-Weinberg equilibrium. Baseline characteristics of cases vs controls were compared using unpaired Student’s t-tests and chi-square tests for continuous and categorical variables, respectively. Chi-square tests were used to test for associations between independent variables and risk of PCOS in univariate analysis. Binary logistic regression was used for multivariate analysis of phenotypic determinants of PCOS risk, with adjustment for factors found to associate with PCOS with P < 0.05 in the univariate analysis of phenotypic determinants. Binary logistic regression was also used to test for association between genotype and risk of PCOS, using an additive model and adjusting for phenotypic factors found to associate independently with PCOS risk (age, body mass index, waist-to-hip ratio, deseasonalized 25(OH)D <10 vs. ≥10 ng/ml, outdoor exercise and fasting glucose). Sub-group analyses were performed to determine whether genetic variation in the vitamin D pathway modified effects of vitamin D status on susceptibility to PCOS by repeating primary efficacy analysis with the inclusion of a term for the interaction between vitamin D status and genotype, using an additive model. Haplotype analysis was performed using the Clark method. Odds ratios are presented with 95% confidence intervals and P values. The Benjamini-Hochberg procedure for multiple testing correction37 was applied to genetic analyses to control the false discovery rate (FDR) at 5%.
Power and sample size
Assuming the risk of vitamin D deficiency (serum 25[OH]D <10 ng/ml) in the control arm to be 56%38, we calculated that 235 cases and 235 controls would need to be recruited in order to detect an odds ratio for the association between vitamin D deficiency and risk of PCOS of ≥1.86 with 80% power and an alpha of 5%.
Data availability
The primary data for this study is available from the corresponding author on direct request.
References
Sirmans, S. M. & Pate, K. A. Epidemiology, diagnosis, and management of polycystic ovary syndrome. Clin. Epidemiol. 6, 1–13 (2013).
Niu, Y.-M. et al. Association Between Vitamin D Receptor Gene Polymorphisms and Polycystic Ovary Syndrome Risk: A Meta-Analysis. Front. Physiol. 9, 1–13 (2019).
Baqai, Z., Khanam, M. & Parveen, S. Prevalence of PCOS in fertile patients. Med. Channel 16, 437–440 (2010).
Rodin, D. A., Bano, G., Bland, J. M., Taylor, K. & Nussey, S. S. Polycystic ovaries and associated metabolic abnormalities in Indian subcontinent Asian women. Clin. Endocrinol. 49, 91–99 (1998).
Diamanti-Kandarakis, E., Kandarakis, H. & Legro, R. S. The role of genes and environment in the etiology of PCOS. Endocr. 30, 19–26 (2006).
Shi, X.-Y., Huang, A.-P., Xie, D.-W. & Yu, X.-L. Association of vitamin D receptor gene variants with polycystic ovary syndrome: a meta-analysis. BMC Med. Genet. 20, 1–11 (2019).
Reis, G. V. O. Pd et al. Vitamin D receptor polymorphisms and the polycystic ovary syndrome: A systematic review. J. Obstet. Gynaecol. Res. 43, 436–446 (2017).
Chen, Y. & Fang, S.-Y. Potential genetic polymorphisms predicting polycystic ovary syndrome. Endocr. Connect. 7, R187–R195 (2018).
He, C., Lin, Z., Robb, S. & Ezeamama, A. Serum vitamin D levels and polycystic ovary syndrome: a systematic review and meta-analysis. Nutrients 7, 4555–4577 (2015).
Krul-Poel, Y. H. M. et al. Vitamin D and metabolic disturbances in polycystic ovary syndrome (PCOS): A cross-sectional study. PLoS one 13, 1–13 (2018).
Krul-Poel, Y. H. M. et al. The role of vitamin D in metabolic disturbances in polycystic ovary syndrome: a systematic review. Eur. J. Endocrinol. 169, 853–865 (2013).
Martineau, A. R. et al. High-dose vitamin D3 during intensive-phase antimicrobial treatment of pulmonary tuberculosis: a double-blind randomised controlled trial. Lancet 377, 242–250 (2011).
Ganmaa, D. et al. High-Dose Vitamin D(3) during Tuberculosis Treatment in Mongolia. A Randomized Controlled Trial. Am. J. Respir. Crit. Care Med. 196, 628–637 (2017).
Martineau, A. R. et al. Association between Gc genotype and susceptibility to TB is dependent on vitamin D status. Eur. Respir. J. 35, 1106–1112 (2010).
Clark, A. G. Inference of haplotypes from PCR-amplified samples of diploid populations. Mol. Biol. Evol. 7, 111–122 (1990).
Mazloomi, S., Sharifi, F., Hajihosseini, R., Kalantari, S. & Mazloomzadeh, S. Association between Hypoadiponectinemia and Low Serum Concentrations of Calcium and Vitamin D in Women with Polycystic Ovary Syndrome. ISRN Endocrinol. 2012, 1–6 (2012).
Hassan, N. E., El-Orabi, H. A., Eid, Y. M. & Mohammed, N. R. Effect of 25-hydroxyvitamin D on metabolic parameters and insulin resistance in patients with polycystic ovarian syndrome. Middle East. Fertil. Soc. J. 17, 176–180 (2012).
Nestler, J. E., Reilly, E. R., Cheang, K. I., Bachmann, L. M. & Downs, R. W. Jr. A pilot study: effects of decreasing serum insulin with diazoxide on vitamin D levels in obese women with polycystic ovary syndrome. Trans. Am. Clin. Climatol. Assoc. 123, 209–220 (2012).
Bacopoulou, F., Kolias, E., Efthymiou, V., Antonopoulos, C. N. & Charmandari, E. Vitamin D predictors in polycystic ovary syndrome: a meta-analysis. Eur. J. Clin. Invest. 47, 746–755 (2017).
Junaid, K., Rehman, A., Jolliffe, D. A., Wood, K. & Martineau, A. R. High prevalence of vitamin D deficiency among women of child-bearing age in Lahore Pakistan, associating with lack of sun exposure and illiteracy. BMC Womens Health 15, 83–83 (2015).
Zakar, M. Z. et al. High-dose vitamin D3 in the treatment of severe acute malnutrition: a multicenter double-blind randomized controlled trial. Am. J. Clin. Nutr. 107, 725–733 (2018).
Roth, D. E. et al. Global prevalence and disease burden of vitamin D deficiency: a roadmap for action in low- and middle-income countries. Ann. N. Y. Acad. Sci. 1430, 44–79 (2018).
Sedighi, S. et al. Comparison of lifestyle in women with polycystic ovary syndrome and healthy women. Glob. J. Health Sci. 7, 228–234 (2014).
Legro, R. S., Dodson, W. C., Kunselman, A. R. & Dunaif, A. Prevalence and Predictors of Risk for Type 2 Diabetes Mellitus and Impaired Glucose Tolerance in Polycystic Ovary Syndrome: A Prospective, Controlled Study in 254 Affected Women1. J. Clin. Endocrinol. Metab. 84, 165–169 (1999).
Azziz, R. & Kashar-Miller, M. Family history as a risk factor for the polycystic ovary syndrome. J. Pediatr. Endocrinol. Metab. 13, 1303–1306 (2000).
Santos, B. R., Lecke, S. B. & Spritzer, P. M. Genetic variant in vitamin D-binding protein is associated with metabolic syndrome and lower 25-hydroxyvitamin D levels in polycystic ovary syndrome: a cross-sectional study. PLoS one 12, 1–12 (2017).
Wehr, E. et al. Vitamin D-associated polymorphisms are related to insulin resistance and vitamin D deficiency in polycystic ovary syndrome. Eur. J. Endocrinol. 164, 741–749 (2011).
Sachs, M. C. et al. Estimating mean annual 25-hydroxyvitamin D concentrations from single measurements: the Multi-Ethnic Study of Atherosclerosis. Am. J. Clin. Nutr. 97, 1243–1251 (2013).
Group, R. E. A.-S. P. C. W. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil. Steril. 81, 19–25 (2004).
PCOS-UK-Guidelines. Diagnosis and management of polycystic ovary syndrome, https://www.guidelines.co.uk/womens-health/pcos-uk-guideline/236071.article (2012).
Khudyakov, P. et al. Prevalence and determinants of QuantiFERON-diagnosed Tuberculosis infection in 9810 Mongolian school children. Clin. Infect. Dis, 813–819 (2018).
Calman, K. Nutrition and bone health: with particular reference to calcium and vitamin D. Rep Health Soc Subj (Lond), 1-24 (1998).
Martineau, A. R. et al. Vitamin D supplementation to prevent acute respiratory tract infections: systematic review and meta-analysis of individual participant data. BMJ 356, 1–14 (2017).
Jolliffe, D. A. et al. Vitamin D supplementation to prevent asthma exacerbations: a systematic review and meta-analysis of individual participant data. Lancet Respir. Med. 5, 881–890 (2017).
Jolliffe, D. A. et al. Vitamin D to prevent exacerbations of COPD: systematic review and meta-analysis of individual participant data from randomised controlled trials. Thorax 74, 337–345 (2019).
Martineau, A. R. Old wine in new bottles: vitamin D in the treatment and prevention of tuberculosis. Proc. Nutr. Soc. 71, 84–89 (2012).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat Methodol. 57, 289–300 (1995).
Hanif, F., Naveed, A. K., Rahim, A. & Waqar, F. Vitamin D level in unmarried females with Polycystic ovarian syndrome. Biochem. 2, 4 (2015).
Acknowledgements
This study was supported by the Higher Education Commission of Pakistan grant number 1-8/HEC/HRD/2017/8369. We thank all participants for taking part in the study. We also thank Shumaila Sadiq and Citilab and Research centre, Lahore, for assistance with participant recruitment and Shahida Hussain for help with laboratory assays.
Author information
Authors and Affiliations
Contributions
Nasira M. Lone designed the study, carried out experimental work and drafted this manuscript. Saba Riaz contributed to study design and Amna Z. Eusaph helped in sample collection and diagnosis of cases. Sidra Younis, David A. Jolliffe, and Kashaf Junaid contributed to the statistical analysis of genetic data. Zhenqiang Wu calculated deseasonalized 25-hydroxyvitamin D levels. Charles A. Mein, Eva L. Wozniak and Theodoros Xenakis helped in genotyping. Adrian R. Martineau contributed to study design, statistical analysis and writing the manuscript. All authors critically reviewed the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lone, N.M., Riaz, S., Eusaph, A.Z. et al. Genotype-independent association between vitamin D deficiency and polycystic ovarian syndrome in Lahore, Pakistan. Sci Rep 10, 2290 (2020). https://doi.org/10.1038/s41598-020-59228-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-020-59228-4
This article is cited by
-
Role of VDR gene polymorphisms and vitamin D levels in normal and overweight patients with PCOS
Journal of Genetics (2024)
-
Heterogeneity in susceptibility to polycystic ovary syndrome among women with epilepsy
Acta Epileptologica (2023)
-
Assessment of Cdx2 polymorphism in Iranian women with polycystic ovary syndrome
Middle East Fertility Society Journal (2023)