Abstract
Chinese fir (Cunninghamia lanceolata) is an important coniferous species that accounts for 20–30% of the total commercial timber production in China. Though traditional breeding of Chinese fir has achieved remarkable success, molecular-assisted breeding has made little progress due to limited availability of genomic information. In this study, a survey of Chinese fir genome was performed using the Illumina HiSeq Xten sequencing platform. K-mer analysis indicated that Chinese fir has a large genome of approximately 11.6 Gb with 74.89% repetitive elements and is highly heterozygous. Meanwhile, its genome size was estimated to be 13.2 Gb using flow cytometry. A total of 778.02 Gb clean reads were assembled into 10,982,272 scaffolds with an N50 of 1.57 kb. In total, 362,193 SSR loci were detected with a frequency of 13.18 kb. Dinucleotide repeats were the most abundant (up to 73.6% of the total SSRs), followed by trinucleotide and tetranucleotide repeats. Forty-six polymorphic pairs were developed, and 298 alleles were successfully amplified from 199 Chinese fir clones. The average PIC value was 0.53, indicating that the identified genomic SSR (gSSR) markers have a high degree of polymorphism. In addition, these breeding resources were divided into three groups, and a limited gene flow existed among these inferred groups.
Similar content being viewed by others
Introduction
Chinese fir (Cunninghamia lanceolata (Lamb.) Hook), an evergreen conifer native to southern China and northern Vietnam, belongs to the Cupressaceae family. Because of its fast growth, desirable wood properties, and high resistant to diseases, it has been widely cultivated in China for over 3000 years. Chinese fir is a timber species that currently has the largest plantation area in China, and its annual harvest accounts for 20–30% of total commercial timber production1. The systematic breeding of Chinese fir, including provenance tests, cross-breeding and clonal selection, has been conducted since the 1960s. Many first-, second- and third-generation seed orchards have been established2. With the accumulation of breeding resources, the management of their genetic diversity and genetic relationships has become increasingly important.
Genetic diversity evaluation of germplasm resources can provide crucial reference information for germplasm conservation, selection of elite germplasms, and parental selection for crosses. In the last twenty years, molecular genetic diversity in Chinese fir has been evaluated many times. You et al. utilized random amplified polymorphic DNA (RAPD) technologies to analyse the genetic diversity of 7 representative provenance samples in China for the first time. They found that there was a relatively high level of genetic diversity among these provenances and deduced that the central-western part of Nanling Mountain is a central distribution zone of Chinese fir3. Subsequently, other molecular markers, including amplified fragment length polymorphism (AFLP)4, sequence-related amplified polymorphism (SRAP)5, and inter simple sequence repeats (ISSRs)6, were also adopted to analyse the molecular genetic diversity of Chinese fir. However, these dominant markers cannot distinguish between heterozygotes and homozygotes; thus, they are insufficient for a precise assessment of molecular genetic diversity in Chinese fir.
Microsatellites or simple sequence repeats (SSRs) are codominant markers and can be divided into expressed sequence tag-SSRs (EST-SSRs) and genomic SSRs (gSSRs). With the advantages of a high level of polymorphism, high specificity and repeatability, and extensive genomic coverage, SSR markers have been widely used to disclose genetic diversity and relationships in many crop species, such as rice (Oryza sativa), wheat (Triticum aestivum), soybean (Glycine max) and Pennisetum species7,8,9,10,11. Few studies on the identification and application of polymorphic SSR markers have been reported in Chinese fir. Using the Chinese fir EST sequences deposited in public databases, Zhang et al. first developed EST-SSR primers and evaluated the molecular genetic diversity of 30 clones12. Through the combination of SSR mining and multiplex-PCR methods, a flow chart of EST-SSR marker development was established using Chinese fir transcriptome data from next-generation sequencing (NGS). In addition, 28 polymorphic EST-SSR loci were obtained, which were verified as suitable for identifying the provenances, even individuals of Chinese fir13. A relatively high level of genetic diversity within different Chinese fir samples was also revealed using EST-SSR primer pairs in previous studies14,15. However, only 11 polymorphic gSSR loci of Chinese fir have been reported thus far, of which 7 derived from candidate genes involved in wood formation, and further experiments showed that the polymorphic information content (PIC) values from gSSR primers were higher than those from EST-SSR primers16,17. In addition, the Chinese fir genome is large, with most sequences being non-coding, and it exhibits abundant variations among different genotypes. Therefore, it is necessary to develop more polymorphic gSSR primers.
NGS has many advantages, including high throughout and rapid sequence data generation. Genome survey sequencing via NGS not only provides information on the genome structure of a species, such as genome size, heterozygosity and repeat contents, but also can generate a large amount of sequence data for the development of new gSSR markers. Recently, many high-quality gSSR markers have been developed in sesame (Sesamum indicum)18, buckwheat (Fagopyrum tataricum)19, pistachio (Pistacia vera)20 and elephant grass (Pennisetum purpureum)21 based on genome survey sequencing. However, until now, there have been no reports on the genome survey sequencing of Chinese fir.
In this study, we aim to (1) estimate the genome size, GC content, and heterozygosity of Chinese fir, (2) conduct a genome-wide identification of SSRs and develop novel gSSR primers, and (3) discover the genetic diversity and population structure of the Chinese fir germplasm. To our knowledge, this is the first report on the genome structure and genome-wide SSRs of Chinese fir. The results will provide valuable information for the whole genome sequencing of Chinese fir and contribute to accelerating the progress of genetic map construction and SSR marker-assisted breeding in this important timber tree.
Results
Genome size estimation by flow cytometry
The nuclei of Chinese fir, barley (Hordeum vulgare), and Populus trichocarpa were stained with propidium iodide (PI). Fluorescent signals were captured using a flow cytometer. The cell peaks of three species are shown in Fig. 1. Most of the nuclei of Chinese fir leaves formed a large peak at 3.67 × 106 (fluorescence intensity) corresponding to the G0/1 phase of the cell cycle (Fig. 1a). Histograms of barley and P. trichocarpa contained two main peaks that corresponded to G0/1 nuclei of both species (Fig. 1b,c). The genome sizes of barley and P. trichocarpa are 5.1 Gb and 480 Mb, respectively22,23. Therefore, the genome size of Chinese fir was calculated to be 13.20 ± 0.57 Gb.
Estimation of genome size in Chinese fir by flow cytometry. G0/1 nuclei peaks (indicated by the arrows) were obtained after flow cytometric analysis of PI-stained nuclei suspensions prepared from leaf samples. Each sample was determined with 10 replicates, and the CV of each peak was set below 5%. (a) G0/1 nuclei peak of Chinese fir (2 C = 26.99 pg, fluorescence intensity 3.67 × 106). (b) G0/1 nuclei peak of barley (2 C = 10.43 pg, fluorescence intensity 1.38 × 106). (c) G0/1 nuclei peak of P. trichocarpa (2 C = 0.98 pg, fluorescence intensity 1.38 × 105).
K-mer analysis
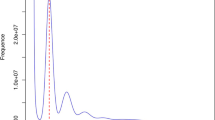
Using the Illumina HiSeq Xten PE150 platform, we obtained 5,292,630,018 clean reads with a total of 778.02 Gb of data, which were used for subsequent K-mer analysis. The 17-mer frequency distribution curve exhibited two peaks at depths of 15 and 30, respectively (Fig. 2). According to the flow cytometry results, it was deducible that the second was the main peak corresponding to the expected K-mer depth. Statistical analysis showed that the total number of K-mers was 347,935,256,501. Using the formula of genome size = total K-mer number/peak depth, the genome size of this sequencing sample was estimated to be 11,597,841,883 bp. The first peak observed at 1/2 of peak depth displayed a high level of heterozygosity for this Chinese fir sample. Simulation analysis using the Arabidopsis thaliana genome revealed that it had a 2.0~2.1% heterozygosity rate (Fig. 2). Similarly, the content of repetitive sequences was estimated to reach 74.89%. In addition, guanine plus cytosine (GC) content analysis under different sequencing depths showed a 36.04% GC content of Chinese fir (Fig. 3).
K-mer (K = 17) analysis for estimating the genome size of Chinese fir. The X-axis is depth (×) and the Y-axis is the proportion that represents the frequency at that depth divided by the total frequency of all depths. The peak indicated by the black arrow is the main peak corresponding to the expected K-mer depth. The genome size is estimated according to the formula: genome size = total K-mer number/peak depth (total K-mer number = 347,935,256,501). Atha. ×39 represents the depth of Arabidopsis thaliana is 39. H0.02 and H0.021 means that heterozygous rate is 2% and 2.1%, respectively. The peaks of Atha. ×39 were used as references.
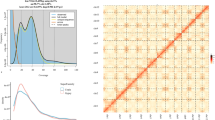
Guanine plus cytosine (GC) content and average sequencing depth of the genome data. The X-axis is the GC content percent across every 10-kb non-overlapping sliding window. The Y-axis represents the average sequencing depth. The distribution of GC content is at the top, while the distribution of sequence depth is on the right side.
Assembly and identification of SSR in Chinese fir
We performed sequence assembly using clean data and obtained 10,982,272 scaffolds with an average length of 693.5 bp. There were 10,982,265 scaffolds longer than 100 bp, and 804,114 were longer than 2 kb (Table 1). The total length of the assembled sequences was 7.62 Gb, exhibiting a relatively large difference from what we estimated (11.6 Gb), suggesting that a systematic sequencing approach must be designed for the large Chinese fir genome with high heterozygosity.
A total of 362,193 SSR loci were identified with the assembled sequences. These loci were distributed on 299,303 scaffolds, with one SSR locus every 13,184 bp (no counting of unknown bases). We grouped these SSR loci according to the number of nucleotides in the repeat motifs (Fig. 4). The dinucleotide motif had the largest number (266,593), accounting for 73.6%, followed by trinucleotide motifs (48,138, 13.3%). The numbers of tetra-, penta- and hexanucleotide motifs were 36,732 (10.1%), 6,791 (1.9%), and 3,939 (1.1%), respectively. The number of SSR-containing sequences decreased as the repeat number of motifs increased. In dinucleotide SSR loci, AT/TA (59.4%) was the most abundant repeat motif, and CG/CG was the least abundant (0.1%) (Fig. 5a). AAT/ATT was the most abundant trinucleotide motif (31.8%), followed by AAG/CTT (21.5%), ATC/ATG (15.9%), AAC/GTT (12.8%) and AGG/CCT (10.3%) (Fig. 5b). The tetra-, penta- and hexanucleotide SSR loci contained more motif types, each in a relatively small percentage (Supplementary Table S1).
Development and screening for polymorphic gSSR markers
In this study, 156 assembled sequences without any unknown base were randomly selected to design the SSR primers, and 156 pairs of primers were successfully designed. Of these primer pairs, 89 were randomly selected to test the successful rate of PCR amplification. Through analysis by 1% agarose gel electrophoresis, 79 (88.76%) SSR primer pairs generated amplification products with expected sizes. The sequences of these 79 SSR loci were deposited into NCBI (accession numbers MK948081- MK959313), and the corresponding repeat motifs and primer sequences were listed in Supplementary Table S2. In order to screen out polymorphic SSR loci, DNA samples of 12 Chinese fir clones from different geographical provenances were used as PCR templates, and capillary electrophoresis of PCR products were further performed to analyse and visualize allelic variation. As an example, three alleles were detected in these 12 clones by CLgSSR1 primers showing three peaks at 160, 163 and 166 bp, respectively (Fig. 6). In total, 46 polymorphic gSSR markers were detected by these primer pairs accounting for 58.2% of effective primers (Table 2).
Allelic variation of gSSR marker CLgSSR1 among 12 Chinese fir clones. To reveal the allelic variation, PCR products of CLgSSR1 primer pairs were analyzed and visualized by capillary electrophoresis. The symbols, such as M33, Cl77, Cl80, etc., on the left of the picture represent different Chinese fir clones. The peaks 1, 2 and 3 represent the 160 bp, 163 bp and 166 bp, respectively.
Diversity survey of novel gSSR loci in Chinese fir
The screened 46 polymorphic gSSR markers were subsequently used to assess their utility in studying genetic variation in 199 Chinese fir clones, and the obtained information included size range, number of alleles (Na), effective alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), Shannon’s information index (I*) and polymorphism information content (PIC) (Table 2).
A total of 298 alleles were detected by the 46 gSSR markers in the 199 clones, and the number of alleles per locus ranged from 2 (CLgSSR28) to 17 (CLgSSR17) with an average of 6.478 alleles per locus. The Ne fluctuated between 1.20 (CLgSSR66) and 6.66 (CLgSSR17) with an average of 2.72 per locus. The locus CLgSSR68 (6.44) and CLgSSR58 (5.62) also showed relatively high Ne values, which were 6.44) and 5.62, respectively. The average of Ho was 0.25, and the Ho value of CLgSSR17 (0.88) was highest among all loci indicating highly heterozygosity of this locus, while the CLgSSR5, CLgSSR7, CLgSSR39 and CLgSSR45 (Ho = 0) were all homozygous in the detected clones. As an important indicator for measuring the genetic diversity of a population, the He of SSRs between 0.3 and 0.8 indicates that a population has higher genetic diversity24. The average He was 0.57, suggesting a high degree of genetic variation for these clones. The Shannon information index (I*) changed between 0.36 (CLgSSR66) and 2.15 (CLgSSR17), and its mean was 1.13. The average value of PIC was 0.53, ranging from 0.18 (CLgSSR66) to 0.83 (CLgSSR17). Based on the classification of Botstein et al.25, 29 loci (63.04%) with PIC values > 0.5 were highly informative, 12 (26.09%) loci were reasonably informative (0.25 < PIC < 0.5), and the others were slightly informative (0 < PIC < 0.25). These results indicated that there was a relatively high level of genetic diversity in this breeding population composed of Chinese fir clones.
Genetic diversity analysis of Chinese fir clones
As mentioned above, all these polymorphic SSR loci were used to estimate the genetic diversity of 199 Chinese fir clones. At first, genetic distance was calculated through pairwise comparisons of the 199 clones by PowerMarker to analyse the genetic similarity between different clones. As a result, the Nei’s genetic distances ranged from 0.0701 to 0.8609 with an average of 0.5073. And most pairwise genetic distances (66.01%) were between 0.4 and 0.6, which indicated high genetic variation among these Chinese fir clones. Moreover, the neighbor-joining (NJ) dendrogram based on Nei’s genetic distances revealed that all clones analysed could be grouped into three major clusters I, II, and III comprised of 70, 37, and 92 clones, respectively (Fig. 7, Supplementary Table S3). On the other hand, to reveal the genetic composition of each clone, we also performed structure analysis based on these 46 polymorphic gSSR loci. According to the method of Evanno et al.26, ΔK was introduced to determine the most appropriate K value, which represents the most likely number of groups. As shown in Fig. 8a, the ΔK value was the highest when K was set to 3. It suggested that these Chinese fir clones were also classified into 3 groups (Fig. 8b). As expected, that most clones were assigned into the same group by clustering of these two methods (Fig. 7, Supplementary Table S3).
Population structure analysis of 199 Chinese fir clones using STRUCTURE software. (a), ΔK was calculated to determine the most appropriate K value for population structure estimation. The modal value of this distribution is the true K (*) or the uppermost level of structure, here three (K = 3) clusters. (b) Population structure of 199 Chinese fir clones. These clones were assigned into three groups (Groups I, II and III) as indicated by the different coloured blocks. Each clone was represented by a single colored line. The greater proportion of a color, the greater the possibility that the represented individual belongs to the group indicated by that color.
According to the report of Wu et al.27, we further analysed the membership probabilities (Q) of Chinese fir clones in the different groups. A total of 184 (92.5%) clones with Q ≥ 0.6 were classified into three groups and were regarded as having a relatively simple genetic structure. Group I had the largest number of clones (80, 40.2%), and it’s worth noting that most of the clones (29/30) from Hunan province were classified into this group. Twenty-five (12.6%) clones belonging to the Groups II were all from Guangxi province. The third group (Group III) contained 79 (39.7%) clones from Rongshui (46), Sanjiang (18), and three other counties (15) of Guizhou province (Supplementary Table S3). The other clones with complicated and mixed origins were placed into the mixed group (Supplementary Table S3). Clones of the same geographical provenance exhibited different genetic compositions. For example, of all clones from Rongshui, Guangxi, 84 were classified into 3 different groups, while the remaining 3 clones belonged to the mixed group. Fifteen clones from Napo, Guangxi were distributed in Groups I and II. Furthermore, some clones from different geographical provenances were classified in the same group suggesting that their genetic background might be similar. Interestingly the mixed group was mainly consisted of 8 clones from Zhejiang and Fujian provinces, which all showed admixture with Groups I and III (Fig. 7). According to the model-based clustering results, an analysis of molecular variance (AMOVA) was conducted and the genetic differentiation coefficient (Fst) was calculated to investigate population differentiation. The results showed that 21.33% (P < 0.001) of the total molecular variation was partitioned among groups, while 38.68% (P < 0.001) of the variation resulted from genetic differences among subgroups and 39.99% (P < 0.001) within subgroups. The pairwise Fst values of the three inferred groups were 0.23 (Group I and II, P < 0.001), 0.19 (Group I and III, P < 0.001) and 0.25 (Group II and III, P < 0.001), respectively. This suggested that these three groups were statistically distinguished from each other. In addition, the gene flow (Nm) was estimated to be 0.92 based on the method of Slatkin et al.28, indicating a limited gene flow among these inferred groups.
Discussion
Genome characteristics of Chinese fir
Based on genome survey sequencing data, we could estimate the genome size of a non-model plant species using K-mer analysis. In the past few years, this approach has been successfully applied to the genomic analysis of some woody plants, such as Chinese bayberry (Myrica rubra)29, Chinese jujube (Ziziphus jujuba)30, and pistachio20. In this study, for the first time, we conducted genome survey sequencing on Chinese fir and obtained 778.02 Gb of clean data. The 17 K-mer and flow cytometry analyses showed that the Chinese fir genome was approximately 11.6–13.2 Gb, which is smaller than the Norway spruce (Picea abies) genome (19.6 Gb)31 and larger than the ginkgo (Ginkgo biloba) genome (10.0 Gb)32. The K-mer analysis also revealed a high level of heterozygosity for the sequencing sample, which was probably due to a high natural outcrossing rate in Chinese fir. The GC content can affect the quality of Illumina sequencing and subsequent assembly33. The GC content of this Chinese fir sample was close to that of wild sweet potato (36%)34 and significantly lower than that of Chinese jujube (48%)30. In addition, the repeat rate was slightly higher than that of elephant grass (71.36%)21. Such information on the Chinese fir genome may provide an important reference for whole-genome sequencing and the selection of assembly strategies in subsequent steps.
Characteristics of gSSR loci in Chinese fir
A total of 362,193 SSR loci were detected from the Chinese fir genome survey sequencing data. The average occurrence frequency of SSR loci was 13.18 kb, which was lower than that in rice (2.75 kb), A. thaliana (2.39 kb), sesame (11.69 kb)18 and pistachio (8.67 kb)20. Among SSR motifs, dinucleotide repeats were the most frequent (73.6%), with AT/AT accounting for 59.4%. Next were the trinucleotide repeats (13.3%), with AAT/ATT accounting for 31.8%. This was similar to the SSR distribution characteristics in the genomes of sesame18, buckwheat19, and pistachio20. However, among the EST-SSRs in Chinese fir, the trinucleotide repeats were the most abundant12,13, suggesting that most of the dinucleotide repeats may be from the intron regions. The massive sequences from the genome survey sequencing provided sufficient data for the identification of high-quality polymorphic SSR loci. The polymorphic SSR markers obtained in this study accounted for 58.2% of the detectable markers, significantly higher than those in Chinese bayberry (31.0%)29 and Metasequoia glyptostroboides (32.9%)35. Of these polymorphic markers, 29 were highly polymorphic (PIC > 0.5).
Genetic diversity and population structure of Chinese fir clones
Polymorphic gSSR markers were used to evaluate the genetic diversity of 199 Chinese fir clones. The average Na, I* and PIC were calculated to be 6.48, 1.125 and 0.526, respectively. Compared to findings from Ouyang and Qi et al.14,36, the breeding resources of Chinese fir used in this study had a higher level of genetic diversity. The model-based population structure analysis could classify the 199 clones into 3 groups, consistent with the results from an NJ method based on Nei’s genetic distance. And, both AMOVA and Fst analyses confirmed the population structure and its statistical significance. The results of the population structure analysis also revealed the existence of introgression and gene flow among different clones. Three groups were identified by two methods, which did not match the clones’ geographical origins. This was in agreement with previous reports regarding Chinese fir37,38, which could be related to wind pollination and a high natural outcrossing frequency in this species, as well as to artificial germplasm exchange. Therefore, the selection of Chinese fir breeding resources should not only focus on geographical origin but also consider the genetic structure of the population.
To gain knowledge on the genome structure of Chinese fir, we had more than 30× Illumina data coverage for its genome survey. The assembled sequences were also used to search for SSR loci, to develop novel gSSR markers, and to study genetic diversity and population structure in this important species. Flow cytometry and K-mer analysis indicated that the Chinese fir genome is 11.6–13.2 Gb in size. The sequencing sample was highly heterozygous and had a high level of repeats, and its GC content was approximately 36.04%. Overall, 362,193 SSR loci were found with a frequency of 13.18 kb. Forty-six polymorphic gSSR primer pairs were developed, and 298 alleles were successfully amplified from 199 Chinese fir clones. These Chinese fir resources could be divided into three groups. In conclusion, in this study, we present the data on the genome structure of the Chinese fir, which may help in designing further whole genome sequencing strategies. Furthermore, we also developed novel polymorphic gSSR markers, which will facilitate germplasm characterization, genetic diversity and population structure studies in this important species.
Materials and Methods
Plant materials and DNA extraction
Chinese fir clone “ZL06” is one of the superior clones with faster growth rate and stronger resistance, and has been widely cultivated as commercial forest in Zhejiang province of China. Due to its commercial value and desirable traits, ZL06 was chosen as material for genome sequencing. Three-year-old trees of this clone was grown in the Pingshan experimental station, Zhejiang A&F University, Hangzhou City, China (30 °15′36.6″N, 119 °42′13.1″E) for genome survey sequencing and flow cytometry analysis. And 199 other Chinese fir clones grown in the Hengzhi germplasm repository (29 °7′24.6″N, 118 °25′57.1″E), Kaihua County, Zhejiang Province, China, were used to test SSR markers for polymorphism and to assess genetic diversity. The numbering and geographical origins of these clones are listed in Supplementary Table S3. The tender leaves, used for DNA isolation, were collected between 21 and 27 of June 2017. After cleaning and disinfection with 70% alcohol, leaves were stored in liquid nitrogen. In addition, the P. trichocarpa and barley tissue culture seedlings used for the flow cytometry analysis were a generous gift from the Zhejiang University Institute of Genetics.
The genomic DNA was extracted from the leaf samples using the CTAB (Cetyltrimethyl- ammonium bromide) method39. DNA purity and concentration were assessed using 1% agarose gel electrophoresis and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). DNA samples were diluted to a concentration of 50 ng/μl for the following SSR-polymerase chain reaction (PCR).
Flow cytometry analysis
The genome size was determined using the CytoFLEX Flow Cytometer (Beckman Coulter, Inc., Brea, CA, USA). A total of 60 mg of fresh leaf tissue was harvested and placed on a Petri dish. One millilitre of lysis buffer (MgSO4 buffer) was added to the Petri dish, and the leaf tissue was cut into pieces with a sharp blade. The mixture was filtered and collected in a 1.5 ml centrifuge tube and centrifuged for 5 min at 1000 rpm/min. The supernatant was discarded, and the pellet was resuspended in 100 μl of pre-chilled lysis buffer. An aliquot of 150 μl of pre-chilled PI staining solution (50 μg/ml) was added for the fluorescence labelling of nuclear DNA. The sample was incubated in the dark at 4 °C for 5 min and then filtered and loaded onto the flow cytometer for detection. The detection for each sample was performed with ten replicates. CyExpert software (Beckman Coulter, Inc.) was used for capturing fluorescent signals and data analysis. Barley (H. vulgare ‘Golden promise’, 2 C = 10.43 pg) and P. trichocarpa (Nisqually-1, 2 C = 0.98 pg) with a known genome size served as reference standards. The relative nuclear DNA content of Chinese fir (C. lanceolate ‘ZL06’) was measured according to o the following formula: Sample 2C-value (pg) = (Sample peak mean/Standard peak mean) * nuclear DNA content of the reference Standard (pg). Genome size was estimated based on the conversion formula: 1 pg DNA = 0.978 × 109 bp40.
Illumina sequencing and K-mer analysis
The genomic DNA of clone ZL06 was randomly fragmented into 350 bp inserts, and a DNA sequencing library was constructed. Then, sequencing (paired-end) was performed at Nextomics Bioscience Co., Ltd. (Wuhan, China). After removing low-quality reads, clean reads were obtained and used for K-mer analysis. The sequence data were deposited in the Genome Sequence Archive41 in BIG Data Center42, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA001775. We used JELLYFISH 2.1.4 to conduct the K-mer analysis and obtained the corresponding frequency distributions43. Based on k-mer frequency distributions, genome size was calculated using the information on peak depth and the number of 17-mer. The heterozygosity rate was also estimated according to k-mer number at half the peak depth and simulation analysis with A. thaliana genome data. The proportion of repeated sequences was analysed using GENOMESCOPE software44.
Sequence assembly and SSR identification
Genome assembly using sequencing data was performed with the SOAPdenovo v2.01 software with the default settings45. For the assembly, contigs were firstly obtained according to the De-Brujin-Graph (DBG) algorithm. After mapping the paired-end reads back to resultant contigs, scaffolds were constructed step by step. Finally, the gaps inside the scaffolds were filled using paired-end reads. SOAPaligner v2.21 was used to analyse the GC-depth distribution46. The scaffolds longer than 100 bp in length were retained. Then, the GC average sequencing depth was calculated by the 10-kb non-overlapping sliding windows along the assembled sequences.
SSR loci were searched using the Perl script MISA (http://pgrc.ipk-gatersleben.de/misa/misa.html). The search parameters were set for the identification of di-, tri-, tetra-, penta-, and hexanucleotide motifs with a minimum number of 6, 5, 4, 4, and 4 repeats, respectively. The numbers of SSR repeats, frequencies of SSRs and motif types were collected and statistically analyzed using Excel 2016 (Microsoft, WA, USA). Web-based BatchPrimer3 v1.0 (http://probes.pw.usda.gov/batchprimer3/) was used for designing SSR primers. The detailed parameters were as follows: 100–300 bp final product length (optimal 150 bp), primer size from 18 to 23 bp (optimal 21 bp) and GC content 40–70% (optimal 50%); the primer melting temperature was set between 50 °C and 70 °C (optimal 55 °C). All primers were synthesized by GenScript Co., Ltd. (Nanjing, China).
PCR and capillary electrophoresis
SSR-PCR was performed in a total volume of 10 μl containing 50 ng DNA, 1× SuperMix containing Taq DNA polymerase (Takara, Dalian, China), and 2 pmol of each primer. The PCR program was as follows: initial denaturation at 94 °C for 5 min, followed by 32 cycles at 94 °C for 30 s, 48~54 °C (determined by primer Tm) for 30 s and 72 °C for 40 s and a final extension at 72 °C for 10 min. Amplified products were first checked using 1% agarose gel electrophoresis. Products with expected sizes were then subjected to capillary electrophoresis. All detections were made in three replicates. SSR alleles were visualized and scored with the Q-Analyzer-1.3.5.1 software (BiOptic Inc.).
Data analysis of genetic diversity and population structure
The 46 polymorphic primer pairs were used to evaluate the genetic diversity of the Chinese fir clones. Parameters including Na, Ne, Ho, He, and I* were analysed with the POPGENE software version 1.3247. The PIC value of each SSR locus was calculated based on the formula25.
where Pi and Pj are the frequency of the ith and jth allele for the given locus, respectively.
Two different approaches were used to assess population structure of Chinese fir clones. The first one was Bayessian clustering analysis implemented with STRUCTURE 2.3.4 software to determine the most likely number of group and assign clones to their most likely group48. The number of groups was set as K (K = 1~15), and each run was replicated 20 times to ensure consistency of results. The burn-in period and Markov chain Monte Carlo (MCMC) were set at 50,000 and 500,000, respectively. The true number of group was determined by the logarithm of likelihood for each K, L(K), and the optimum value ΔK was obtained by the formula26.
where s[L(K)] is the standard deviation of L(K). The Q data of the 20 replicate runs of the best value of K were integrated by CLUMPP software49. The second clustering was based on Nei’s 1983 genetic distance and on the NJ algorithm using PowerMarker v3.25 software50. This bootstrap analysis was carried out with 1,000 replicates. And MEGA 7.0 software was used to plot the dendrogram51. In addition, to summarize the major patterns of variation within the multi-locus dataset, an AMOVA analysis was performed using GenAlEx V6.5 with 999 permutations52. Nm of whole population was also estimated using the formula28.
Data availability
The sequence data reported in this study have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA001775, CRA001775 that are publicly accessible at http://bigd.big.ac.cn/gsa.
References
Huang, H. H. et al. De novo characterization of the Chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genomics 13 (2012).
Li, M., Chen, X., Huang, M., Wu, P. & Ma, X. Genetic diversity and relationships of ancient Chinese fir (Cunninghamia lanceolata) genotypes revealed by sequence-related amplified polymorphism markers. Genetic Resources and Crop Evolution 64, 1087–1099 (2017).
Yong, Y. & Hong, J. S. Application of RAPD marker to genetic variation of Chinese fir provenances. Scientia Silvae Sinicae 34, 32–38 (1998).
Chung, J. D., Lin, T. P., Tan, Y. C., Lin, M. Y. & Hwang, S. Y. Genetic diversity and biogeography of Cunninghamia konishii (Cupressaceae), an island species in Taiwan: a comparison with Cunninghamia lanceolata, a mainland species in China. Molecular Phylogenetics and Evolution 33, 791–801 (2004).
Zheng, H. Q., Hu, D. H., Wang, R. H. & Wu, S. J. Genetic divergence of the Chinese fir fast-growing genotypes revealed by SRAP markers. Journal of Southwest Forestry University 37, 14–20 (2017).
Chen, Y. et al. Genetic diversity and variation of Chinese fir from Fujian province and Taiwan, China, based on ISSR markers. PloS one 12, e0175571 (2017).
Yu, J. K., Rota, M. L., Kantety, R. V. & Sorrells, M. E. EST derived SSR markers for comparative mapping in wheat and rice. Molecular Genetics and Genomics 271, 742–751 (2004).
Wang, L. X., Guan, R. X., Liu, Z. X., Chang, R. Z. & Qiu, L. J. Genetic diversity of Chinese cultivated soybean revealed by SSR markers. Crop Science 46, 1032–1038 (2006).
Aljumaili, S. J. et al. Genetic diversity of aromatic rice germplasm revealed by SSR markers. BioMed Research International 2018, 1–11 (2018).
Zhou, S. et al. Phylogenetics and diversity analysis of Pennisetum species using Hemarthria EST-SSR markers. Grassland Science 65, 13–22 (2019).
Zhou, S. et al. The first Illumina-based de novo transcriptome analysis and molecular marker development in Napier grass (Pennisetum purpureum). Molecular Breeding 38, 95 (2018).
Zhang, S., Huang, H. H., Lin, E. P. & Tong, Z. K. Development and application of EST-SSR markers for Cunninghamia lanceolate and Taiwania cryptomerioides. Scientia Silvae Sinicae 49 (2013).
Wen, Y. F., Han, W. J., Zhou, H. & Xu, G. B. SSR mining and development of EST-SSR markers for Cunninghamia lanceolata based on transcriptome sequences. Scientia Silvae Sinicae 51, 40–49 (2015).
Ouyang, L. et al. Genetic diversity among the germplasm collections of the Chinese fir in 1st breeding population upon SSR markers. Journal of Nanjing Forestry University (Natural Sciences Edition) 38, 21–26 (2014).
Duan, H. et al. Genetic characterization of Chinese fir from six provinces in couthern China and construction of a core collection. Scientific Reports 7, 13814 (2017).
Xu, Y. et al. Development of EST-SSR and genomic-SSR in Chinese fir. Journal of Nanjing Forestry University (Natural Sciences Edition) 38, 9–14 (2014).
Han, Y. et al. Development and application of SSR loci from functional genes involved in wood formation in Cunninghamia lanceolata. Journal of Agricultural Biotechnology 27, 38–46 (2019).
Wei, X. et al. Development of simple sequence repeat (SSR) markers of sesame (Sesamum indicum) from a genome survey. Molecules 19, 5150–5162 (2014).
Hou, S. Y. et al. Genetic diversity of buckwheat cultivars (Fagopyrum tartaricum Gaertn.) assessed with SSR markers developed from genome survey sequences. Plant Molecular Biology Reporter 34, 233–241 (2016).
Motalebipour, E. Z., Kafkas, S., Khodaeiaminjan, M., Çoban, N. & Gözel, H. Genome survey of pistachio (Pistacia vera L.) by next generation sequencing: Development of novel SSR markers and genetic diversity in Pistacia species. BMC Genomics 17, 998 (2016).
Wang, C. et al. Genome survey sequencing of purple elephant grass (Pennisetum purpureum Schum ‘Zise’) and identification of its SSR markers. Molecular Breeding 38, 94 (2018).
Consortium, T. I. B. G. S. et al. A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716 (2012).
Tuskan, G. A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (2006).
Nei, M. & Takezaki, N. The root of the phylogenetic tree of human populations. Molecular Biology and Evolution 13, 170–177 (1996).
Botstein, D., White, R. L., Skolnick, M. & Davis, R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics 32, 314–331 (1980).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14, 2611–2620 (2005).
Wu, K. et al. Genetic analysis and molecular characterization of Chinese sesame (Sesamum indicum L.) cultivars using insertion-deletion (InDel) and simple sequence repeat (SSR) markers. BMC Genetics 15, 35 (2014).
Slatkin, M. & Barton, N. H. A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43, 1349–1368 (1989).
Jiao, Y. et al. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra). BMC Genomics 13, 201–216 (2012).
Xiao, J. et al. Genome-wide characterization of simple sequence repeat (SSR) loci in Chinese jujube and jujube SSR primer transferability. PLoS ONE 10, 1–13 (2015).
Nystedt, B. et al. The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584 (2013).
Guan, R. et al. Draft genome of the living fossil Ginkgo biloba. GigaScience 5, 49 (2016).
Aird, D. et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biology 12, R18 (2011).
Hirakawa, H. et al. Survey of genome sequences in a wild sweet potato, Ipomoea trifida (H. B. K.) G. Don. DNA Research 22, 171–179 (2015).
Zhang, X. Y., Zhang, Y. D., Peng, C., Song, C. W. & Yang, Y. L. Analysis of genomic microsatellite sequence and development of SSR markers in Metasequoia glyptostroboides. Scientia Silvae Sinicae 49, 160–166 (2013).
Qi, M. Genetic diversity of wide cross population of Cunninghamia lanceolata and Platycladu orientalis. Bulletin of Botanical. Research 28, 299–303 (2008).
Chen, Y. Q. et al. Analysis of genetic relationship among Chinese fir (Cunninghamia lanceolata hook) provenances by RAPD. Chinese Journal of Applied & Environmental Biology 7, 130–133 (2001).
Duan, H. J. et al. Genetic characterization of red-colored heartwood genotypes of Chinese fir using simple sequence repeat (SSR) markers. Genetics Molecular Research 14, 18552–18561 (2015).
Porebski, S., Bailey, L. G. & Baum, B. R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter 15, 8–15 (1997).
Doležel, J. & Bartoš, J. Plant DNA flow cytometry and estimation of nuclear genome size. Annals of botany 95, 99–110 (2005).
Wang, Y. et al. GSA: genome sequence archive*. Genomics, Proteomics & Bioinformatics 15, 14–18 (2017).
Members, B. D. C. Database resources of the BIG data center in 2019. Nucleic Acids Research 47, D8–D14 (2018).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Vurture, G. W. et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204 (2017).
Luo, R. B. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 1–6 (2012).
Li, R. Q. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
Yeh, F. C., Yang, R., Boyle, T., Ye, Z. & Mao, J. X. POPGENE, version 1.32: the user friendly software for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton, AB, Canada (1999).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Jakobsson, M. & Rosenberg, N. A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007).
Liu, K. J. & Muse, S. V. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129 (2005).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874 (2016).
Peakall, R. & Smouse, P. E. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6, 288–295 (2006).
Acknowledgements
We thank the anonymous referees and the editors for their comments and suggestions for improving the manuscript. This work was supported in part by the National Key Research and Development Program of China (Grant no. 2017YFD0600201), the Tree Breeding Research Project of Zhejiang Province (2016C02056-5), and the State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University) (201201).
Author information
Authors and Affiliations
Contributions
H.H. and Z.T. conceived and designed this work. E.L. and H.Z. conducted the bioinformatic analysis. H.Z. and J.Y. performed the SSR-PCR and population structure analysis. X.L. and M.Z. performed the flow cytometry analysis. E.L. and H.H. prepared the manuscript. All the authors have read and approved the publication of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lin, E., Zhuang, H., Yu, J. et al. Genome survey of Chinese fir (Cunninghamia lanceolata): Identification of genomic SSRs and demonstration of their utility in genetic diversity analysis. Sci Rep 10, 4698 (2020). https://doi.org/10.1038/s41598-020-61611-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-020-61611-0
This article is cited by
-
Genome survey and identification of simple sequence repeat markers for Caucasian clover (Trifolium ambiguum Bieb.)
Genetic Resources and Crop Evolution (2025)
-
Genetic diversity and population structure analysis in a large collection of Vicia amoena in China with newly developed SSR markers
BMC Plant Biology (2021)
-
Influence of juvenile and mature wood on anatomical and chemical properties of early and late wood from Chinese fir plantation
Journal of Wood Science (2021)
-
Genome-wide analysis and functional characterization of the DELLA gene family associated with stress tolerance in B. napus
BMC Plant Biology (2021)
-
Comparative genomic study on the complete plastomes of four officinal Ardisia species in China
Scientific Reports (2021)