Abstract
Melanized fungi and black yeasts in the family Herpotrichiellaceae (order Chaetothyriales) are important agents of human and animal infectious diseases such as chromoblastomycosis and phaeohyphomycosis. The oligotrophic nature of these fungi enables them to survive in adverse environments where common saprobes are absent. Due to their slow growth, they lose competition with common saprobes, and therefore isolation studies yielded low frequencies of clinically relevant species in environmental habitats from which humans are thought to be infected. This problem can be solved with metagenomic techniques which allow recognition of microorganisms independent from culture. The present study aimed to identify species of the family Herpotrichiellaceae that are known to occur in Brazil by the use of molecular markers to screen public environmental metagenomic datasets from Brazil available in the Sequence Read Archive (SRA). Species characterization was performed with the BLAST comparison of previously described barcodes and padlock probe sequences. A total of 18,329 sequences was collected comprising the genera Cladophialophora, Exophiala, Fonsecaea, Rhinocladiella and Veronaea, with a focus on species related to the chromoblastomycosis. The data obtained in this study demonstrated presence of these opportunists in the investigated datasets. The used techniques contribute to our understanding of environmental occurrence and epidemiology of black fungi.
Similar content being viewed by others
Introduction
A large number of species of black yeast-like fungi that belong to the ascomycetous order Chaetothyriales in the family Herpotrichiellaceae are renowned as opportunistic pathogens in immunocompetent vertebrate hosts1,2. Agents are particularly involved in subcutaneous, and systemic or disseminated infections, known as chromoblastomycosis and phaeohyphomycosis, respectively2,3,4. These infections are invariably chronic and can be severely mutilating or even fatal.
Chromoblastomycosis is a relatively common disease in rural tropical climate zones around the world. This implantation disorder is characterized by the presence of a specialized tissue form of the fungus known as the muriform cell2,5,6. Infection is hypothesized to take place via traumatic inoculation of environmental material such as plant thorns and/or wood fragments7,8. Epidemiological data confirmed by studies using selective isolation methods9,10,11,12 suggest an environmental origin of this disease. However, presence of these agents is infrequent. Only few isolates have been recovered even after extensive sampling in endemic areas9, 10,13,14, where cultures usually only yield non-pathogenic relatives. Novel molecular methods are required for understanding the ecology and environmental occurrence of these agents.
Metagenomics are culture-independent methods for the study of microbial diversity, based on next generation sequencing (NGS) and allowing characterization of fungi in complex environmental systems, using specific molecular markers for identification15. Abundant metagenomic data are available in public databases such as Sequence Read Archive (SRA16), Rast Server (MG-RAST17), and EBI metagenomics (EMG18). Likewise, sequences of several molecular markers are available that are in use for taxonomy and routine molecular identification of species in Herpotrichiellaceae, i.e. ITS, TEF1, BT2, and ACT119. Alternatively, padlock probes, which are specific oligonucleotides with the ability to identify single nucleotide polymorphisms (SNPs), have been proposed for the recognition of several groups of black agents20,21,22,23,24,25. DNA barcoding, based on the ITS region and applying short sequences (25‒41 bp) of nucleotides specific for a single taxonomic species26, can additionally be used to recognize herpotrichiellaceous species by variable regions in the ribosomal operon.
The present study aims to explore the environmental occurrence of chromoblastomycosis agents in the family Herpotrichiellaceae in environmental samples in tropical areas of Brazil. We compare metagenomic data present in public databases, using barcodes and padlock probes for species identification. This approach should lead to better understanding of the sources and routes of infection of patients with chromoblastomycosis.
Results
Datasets containing herpotrichiellaceous fungi
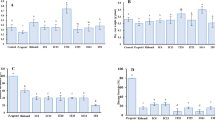
In total, 169 large datasets distributed in 3,786 samples from Brazil were analyzed (Table S1). Of these, only 11 large datasets arranged in 179 samples have sequences of members of Herpotrichiellaceae, originating from five states and representing environmental samples from different geographic areas (Fig. 1A).
Herpotrichiellaceous sequences encountered in investigated datasets. (A) Geographic metagenomic data distribution. (B) Total of reads in investigated datasets. (C) Herpotrichellaceous sequences per dataset. The image was created using Adobe Photoshop CC (v. 20.0.4) based on the map (https://commons.wikimedia.org/wiki/File:20111110231441!Estados_de_nascimento_de_presidentes_brasileiros.png), which is available under a Creative Commons license https://creativecommons.org/licenses/by-sa/3.0/deed.en.
The generated data was according to the scope of each metagenome project evaluated, which resulted in a high variation in size of the datasets. The read number ranged from 14,293 to 1,394,769,476, with the rhizosphere metadata (PRJNA362455) being the one with the highest number of reads (Table 1; Fig. 1B). Within each read pool, the ones matching Herpotrichiellaceae ranged from 4 reads to 14,821 sequences, with the highest concentration in the plant metadata (PRJNA522264). All results considered normalized data (Table 1; Fig. 1C).
The total number of reads matching herpotrichiellaceous fungi was 18,329. Of this data pool, 84% (15,526 reads) were identified by barcode markers, and only around 5.6% (1,032 reads) exclusively by padlock probe markers. The number of sequences identified simultaneously by both markers were 1,771 reads (Table 1), which underlined the requirement to use more than a single tool for in silico identification.
Species identified
In the datasets investigated, the genera Cladophialophora, Exophiala, Fonsecaea, Rhinocladiella and Veronaea were identified. The sequences mainly belonged to the genus Exophiala, which was identified by barcodes and padlock probes. Among the fungi from the family Herpotrichiellaceae, Exophiala species were the most abundantly represented (18,113 sequences) corresponding to 98.77% of the total sequences belonging to 16 described Exophiala species of which E. bergeri (46.01%), E. sideris (27.86%), and E. pisciphila (11.42%) were prevalent. The presence of Fonsecaea pedrosoi, the major agent of chromoblastomycosis in Brazil, was detected at low incidence (0.74%). Cladophialophora species (0.14%) were represented by C. chaetospira (0.12%), and C. arxii and C. immunda, both with 0.01%. Of the genus Rhinocladiella (0.30%), two species were identified, i.e. R. similis (0.08%) and R. atrovirens (0.22%). Veronaea botryosa was present in low numbers (0.05%) (Table 2).
Among the datasets analyzed, the largest number of species was found in soil-associated material and in plants of the family Velloziaceae (PRJNA522264) and in root-associated debris of maize (PRJEB24131), i.e. E. sideris, E. xenobiotica, E. oligosperma, F. pedrosoi and R. atrovirens which were found only in the first dataset, and E. spinifera, E. pisciphila, E. equina, R. similis and V. botryosa in second dataset, while E. bergeri, E. brunnea and E. cancerae were present in both sources. In maize rhizosphere (PRJNA379918) and citrus rhizosphere (PRJNA362455) E. pisciphila and E. xenobiotica were found, while E. brunnea was present only in the citrus source and E. exophialae in maize. In mycorrhizal fungi (PRJNA339563), E. castellanii, E. oligosperma, E. brunnea. E. xenobiotica were identified. In mangrove (PRJNA478407), E. angulospora, E. pisciphila, C. arxii, C. chaetospira and V. botryosa were present. In lignocellulosic biomass (PRJEB5245), E. jeanselmei and E. cancerae were identified. In soils contaminated with crude oil (PRJNA421085), E. pisciphila, C. immunda and C. chaetospira were present. Moreover, in the sugarcane filter cake (PRJNA285006), E. dermatitidis, E. mesophila, E. spinifera, E. bergeri, E. cancerae, E. xenobiotica and V. botryosa were identified. The river water source (PRJNA237344) showed sequences of E. heteromorpha, E. dermatitidis and E. exophialae. The dataset with sequences associated with ants (PRJNA321130) identified only E. exophialae (Table 2).
Discussion
In this study we investigated the presence of sequences of herpotrichiellaceous fungi in metagenomic datasets that were generated after analysis of divergent environmental sources, using molecular markers for in silico identification of causal agents of chromoblastomycosis and phaeohyphomycosis. The tools used as reference were padlock probes developed for rapid detection of pathogenic Fonsecaea species in clinical samples (F. pedrosoi, F. nubica, F. monophora and F. pugnacius20,24), the agent of neurotropic phaeohyphomycosis Cladophialophora bantiana23, and other opportunistic species with variable pathology21,22,25. ITS rDNA barcoding sequences had previously been recommended for rapid identification of clinical and environmental sequences27, and were suggested for taxonomic identification in metagenomic data26.
The results indicated that this methodology represents complementary data to studies on direct isolation via culture9,10,11,12,13,14,28, which all reported low frequency of these agents in the environment. Judging from the number of sequences present in the evaluated datasets, the low frequency of herpotrichiellaceous fungi, compared to the total number of fungal sequences, was confirmed (Table 1). For example, Fonsecaea pedrosoi, a major agent of chromoblastomycosis in Brazil2, was detected in metagenomic data from plant- and soil-associated materials. This habitat is in line with the hypothesis of chromoblastomycosis as an implantation disease from inoculated plant-derived material. This demonstrates that in silico identification can be used as a new tool to uncover the natural habitat of agents of opportunistic diseases and assists in elucidating the environmental occurrence and the route of infection of causative species.
The infection route of agents of chromoblastomycosis nevertheless remains controversial. Their occurrence in living plants has extensively been discussed. Previous studies have shown that Fonsecaea species occurring in living plant material mostly belong to other species than those repeatedly encountered on the human host13,29. In our study, the non-pathogenic Fonsecaea species were not detected. A study presented an in vitro plant infection model showing that the agents of human chromoblastomycosis have a certain degree of plant-invasive ability30, suggesting that those species occur on plants as well. We may hypothesize, that both strictly saprobic and opportunistic species are very rare and thus both have a low chance to be detected in non-optimal datasets using unbiased methodology. Differences in habitat choice, even when minute, may influence species-specific population dynamics and representation in metagenomics datasets, slight differences determining presence or absence.
Species of the genus Rhinocladiella have been described as less common agents of chromoblastomycosis31,32, i.e. R. aquaspersa, R. similis and R. tropicalis3. The extremely rare agent Rhinocladiella similis has also been isolated from dialysis water and from babassu coconuts14,33, while in our in silico data, R. similis was observed in the rhizosphere of maize. The human host thus is unlikely to be the prime habitat of R. similis. The saprobe R. atrovirens was identified in plant and soil-associated habitats. In addition, Veronaea botryosa, an extremely rare agent of disseminated infections in patients with CARD9 immune disorders34,35, had previously been isolated from babassu coconuts14 and from creosote-treated railway ties10. In this study, the species was identified in mangrove, maize rhizosphere and in sugarcane filter cake, indicating a wider saprobic occurrence.
Presence of herpotrichiellaceous opportunists in the environment has been shown by several authors8,9,10,11,12,13,14,28. Our in silico data showed that the most common sequences in metagenomic databases belonged to the genus Exophiala. This is the largest genus in the family Herpotrichiellaceae containing numerous species, many of which are opportunistic pathogens of cold- and warm-blooded animals19,36. We detected species reported from various types of disease other than chromoblastomycosis, i.e. E. bergeri, E. dermatitidis, E. jeanselmei, E. heteromorpha, E. mesophila, E. spinifera, E. oligosperma and E. xenobiotica37. Also E. angulospora, E. pisciphila and E. equina, associated with infections of cold-blooded animal such as frogs, toads and fish36,38 were detected. Exophiala cancerae was first described from the Lethargic crab disease (LCD) occurring along the Brazilian coas36,39. This species hitherto had only been found in endemic coastal areas. However, in our study it was identified in soil, plant roots and in a sugar filter cake, indicating a wider environmental occurrence. Other unexpected encounters were E. castellanii, previously isolated from water40 but in our data among mycorrhizal fungi, E. brunnea, known from litter36 but here in association with mycorrhizal fungi, rhizosphere and plant, and E. sideris from the hydrocarbon-polluted environments41 but here from plant- and soil-associated materials, and finally E. exophialae known from straw in a burrow of Dasypus septemcinctus, but here from river water, rhizosphere and associated with ants.
The genus Cladophialophora was represented by two opportunistic species, C. arxii and C. immunda. Cladophialophora arxii was originally reported from a disseminated infection42 and C. immunda from a patient with a subcutaneous ulcer43. The latter species was later detected in sites polluted with hydrocarbons44, which matches with its presence in soils contaminated with crude oil analyzed in this study. The environmental saprobe C. chaetospira is known to occur in plant litter10, 43, while in our study it was found in mangroves and in soil contaminated with crude oil.
Conclusions
The methodology presented in this study was shown to be a reliable and quick alternative to identify the presence of agents of clinical interest in environmental samples, which is particularly valid for fungi that are difficult to bring in culture, such as black yeasts and other opportunistic agents of human disease. The use of molecular markers as tools for the identification of Herpotrichiellaceae in metagenomic datasets proved to be an effective way to study microhabitats of these fungi, demonstrating the importance of mining databanks for tracking fungal agents. Although local, Brazilian databases were used, the investigated fungi have global distributions, and results are likely to be similar elsewhere. However, data availability is still limited, since the barcode sequences and padlocks described in the literature are restricted to relatively few species. This may explain why in a number of cases our data are significantly different from existing literature, in that common saprobic relatives were not detected, while species with supposedly limited distribution were found in remote, variable habitats suggesting a low degree of host- or habitat-specificity. Expansion of databases may provide a more balanced picture in the future.
Materials and methods
Database construction
The metagenomic database was created based on projects disponible in the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra). To search the projects, the term “metagenomic Brazil” was used and all projects were downloaded. This dataset contained a total of 3,786 samples with approximately 2 terabytes (Table S1). The database was assembled only with metagenomes that complied with four criteria: (1) DNA sequences; (2) Brazilian projects to narrow down the selection; (3) environmental link (arthropods and other animals, aquatic bodies, hostile environments including rocks, decomposing materials with plant debris and soil), since within the geographic area the actual habitat is unknown; (4) public data available for download in the SRA. The datasets were rearranged according to eight types of sources, i.e. rhizosphere (PRJNA379918, PRJNA362455, PRJEB24131), ant (PRJNA321130), aquatic (PRJNA237344), biotechnological (PRJNA285006, PRJEB5245), mycorrhizal (PRJNA339563), plant (PRJNA522264), mangrove (PRJNA478407), and soil (PRJNA421085) (Table 3).
Identification tools
The molecular markers for members of the family Herpotrichiellaceae described in the literature (Table S2) were used for species identification in the metagenome datasets. A total of 97 barcode identifiers with 25‒41 bp26 and 25 padlock probes sequences with 28‒42 bp with different SNPs were collected from an rDNA internal transcribed spacer (ITS220,21,22,23,24,25).
Identification in silico
Comparison of metagenomes with molecular marker sequences was performed with local BLASTn (v2.6.0.+). For the data mining, only alignments with coverage and identity cutoff of 100% (perfect match) were considered (Fig. 2). Matches with values below the cutoff were excluded. Because padlock and barcode probes are extremely specific for species identification, cases of slight misalignment and non-perfect sequence identity do not characterize the fungus in the analyses (Fig. 2). Metagenome reads from double-strand sequencing where considered once in the final read count.
References
Deng, S. et al. Global spread of human chromoblastomycosis is driven by recombinant Cladophialophora carrionii and predominantly clonal Fonsecaea species. PLoS Negl. Trop. Dis. 9, 1–15. https://doi.org/10.1371/journal.pntd.0004004 (2015).
Queiroz-Telles, F. et al. Chromoblastomycosis. Clin. Microbiol. Rev. 30, 233–276. https://doi.org/10.1128/CMR.00032-16 (2017).
Gomes, R. R. et al. Molecular epidemiology of agents of human chromoblastomycosis in brazil with the description of two novel species. PLoS Negl. Trop. Dis. 10, 1–20. https://doi.org/10.1371/journal.pntd.0005102 (2016).
Kirchhoff, L., Olsowski, M., Rath, P. M. & Steinmann, J. Exophiala dermatitidis: Key issues of an opportunistic fungal pathogen. Virulence 10, 1–15. https://doi.org/10.1080/21505594.2019.1596504 (2019).
Seyedmousavi, S. et al. Black yeasts and their filamentous relatives: Principles of pathogenesis and host defense. Clin. Microbiol. Rev. 27, 527–542. https://doi.org/10.1128/CMR.00093-13 (2014).
De Azevedo, C. M. P. S. et al. Fonsecaea pugnacius, a novel agent of disseminated chromoblastomycosis. J. Clin. Microbiol. 53, 2674–2685. https://doi.org/10.1128/JCM.00637-15 (2015).
Marques, S. G. et al. Isolation of Fonsecaea pedrosoi from the shell of the babassu coconut (Orbignya phalerata Martius) in the Amazon region of Maranhão Brazil. Jpn. J. Med. Mycol. 47, 305–311. https://doi.org/10.3314/jjmm.47.305 (2006).
Salgado, C. G. et al. Isolation of Fonsecaea pedrosoi from thorns of Mimosa pudica, a probable natural source of chromoblastomycosis. Rev. Inst. Med. Trop. Sao Paulo 46, 33–36. https://doi.org/10.1590/S0036-46652004000100006 (2004).
Vicente, V. A., de Angelis, D. A., Queiróz-Telles Filho, F. & Pizzirani-Kleiner, A. A. Isolation of herpotrichiellacious fungi from the environment. Brazil. J. Microbiol. 32, 47–51. https://doi.org/10.1590/S1517-83822001000100011 (2001).
Vicente, V. A. et al. Environmental isolation of black yeast-like fungi involved in human infection. Stud. Mycol. 61, 137–144. https://doi.org/10.3114/sim.2008.61.14 (2008).
Guerra, R. S. et al. Black yeast biota in the mangrove, in search of the origin of the lethargic crab disease (LCD). Mycopathologia 175, 421–430. https://doi.org/10.1007/s11046-013-9636-1 (2013).
Satow, M. M., Attili-Angelis, D., de Hoog, G. S., Angelis, D. F. & Vicente, V. A. Selective factors involved in oil flotation isolation of black yeasts from the environment. Stud. Mycol. 61, 157–163. https://doi.org/10.3114/sim.2008.61.16 (2008).
Vicente, V. A. et al. Environmental siblings of black agents of human chromoblastomycosis. Fungal Divers. 65, 47–63. https://doi.org/10.1007/s13225-013-0246-5 (2014).
Nascimento, M. M. F. et al. Diversity of opportunistic black fungi on babassu coconut shells, a rich source of esters and hydrocarbons. Fungal Biol. 121, 488–500. https://doi.org/10.1016/j.funbio.2017.01.006 (2017).
Orellana, S. C. et al. Assessment of fungal diversity in the environment using metagenomics: A decade in review. Fungal Genomics Biol. 03, 110. https://doi.org/10.4172/2165-8056.1000110 (2013).
Leinonen, R., Sugawara, H. & Shumway, M. The sequence read archive. Nucleic Acids Res. 39, 2010–2012. https://doi.org/10.1093/nar/gkq1019 (2011).
Glass, E. M. & Meyer, F. The metagenomics rast server: A public resource for the automatic phylogenetic and functional analysis of metagenomes. In Handbook of Molecular Microbial Ecology I: Metagenomics and Complementary Approaches (ed. de Bruijn, F. J.) 325–331 (Wiley-Blackwell, 2011).
Mitchell, A. et al. EBI metagenomics in 2016—An expanding and evolving resource for the analysis and archiving of metagenomic data. Nucleic Acids Res. 44, D595–D603. https://doi.org/10.1093/nar/gkv1195 (2016).
Teixeira, M. M. et al. Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales, Ascomycota). Stud. Mycol. 86, 1–28. https://doi.org/10.1016/j.simyco.2017.01.001 (2017).
Najafzadeh, M. J., Sun, J., Vicente, V. A. & de Hoog, G. S. Rapid identification of fungal pathogens by rolling circle amplification using Fonsecaea as a model. Mycoses 54, 577–582. https://doi.org/10.1111/j.1439-0507.2010.01995.x (2011).
Najafzadeh, M. J. et al. Detection and identification of opportunistic Exophiala species using the rolling circle amplification of ribosomal internal transcribed spacers. J. Microbiol. Methods. 94, 338–342. https://doi.org/10.1016/j.mimet.2013.06.026 (2013).
Najafzadeh, M. J. et al. Rapid identification of seven waterborne Exophiala species by RCA DNA padlock probes. Mycopathologia 183, 669–677. https://doi.org/10.1007/s11046-018-0256-7 (2018).
Hamzehei, H. et al. Use of rolling circle amplification to rapidly identify species of Cladophialophora potentially causing human infection. Mycopathologia 175, 431–438. https://doi.org/10.1007/s11046-013-9630-7 (2013).
Schneider, G. X. et al. New molecular markers distinguishing Fonsecaea agents of chromoblastomycosis. Mycopathologia 184, 493–504. https://doi.org/10.1007/s11046-019-00359-2 (2019).
Deng, S. et al. Three isothermal amplification techniques for rapid identification of Cladophialophora carrionii, an agent of human chromoblastomycosis. J. Clin. Microbiol. 52, 3531–3535. https://doi.org/10.1128/JCM.01033-14 (2014).
Heinrichs, G., De Hoog, G. S. & Haase, G. Barcode identifiers as a practical tool for reliable species assignment of medically important black yeast species. J. Clin. Microbiol. 50, 3023–3030. https://doi.org/10.1128/JCM.00574-12 (2012).
Zeng, J. S. & de Hoog, G. S. Exophiala spinifera and its allies: Diagnostics from morphology to DNA barcoding. Med. Mycol. 46, 193–208. https://doi.org/10.1080/13693780701799217 (2008).
Feng, P. Y. et al. Cladophialophora abundans, a novel species of Chaetothyriales isolated from the natural environment. Mycol. Prog. 13, 381–391. https://doi.org/10.1007/s11557-013-0924-4 (2014).
De Hoog, G. S. et al. Molecular analysis and pathogenicity of the Cladophialophora carrionii complex, with the description of a novel species. Stud. Mycol. 58, 219–234. https://doi.org/10.3114/sim.2007.58.08 (2007).
Fornari, G. et al. A model for trans-kingdom pathogenicity in Fonsecaea agents of human chromoblastomycosis. Front. Microbiol. 9, 1–11. https://doi.org/10.3389/fmicb.2018.02211 (2018).
González, G. M., Rojas, O. C., González, J. G., Kang, Y. & De Hoog, G. S. Chromoblastomycosis caused by Rhinocladiella aquaspersa. Med. Mycol. Case Rep. 2, 148–151. https://doi.org/10.1016/j.abd.2019.09.014 (2013).
Richarz, N. A. et al. First case of chronic cutaneous chromoblastomycosis by Rhinocladiella similis aquired in Europe. Clin. Exp. Dermatol. 43, 925–927. https://doi.org/10.1111/ced.13659 (2018).
Figel, I. C. et al. Black yeasts-like fungi isolated from dialysis water in hemodialysis units. Mycopathologia 175, 413–420. https://doi.org/10.1007/s11046-013-9633-4 (2013).
Bonifaz, A. et al. Severe disseminated phaeohyphomycosis in an immunocompetent patient caused by Veronaea botryosa. Mycopathologia 175, 497–503. https://doi.org/10.1007/s11046-013-9632-5 (2013).
Welfringer, A. et al. A rare fungal infection: Phaehyphomycosis due to Veronaea botryosa and review of literature. Med. Mycol. Case Rep. 15, 21–24. https://doi.org/10.1016/j.mmcr.2017.02.001 (2017).
de Hoog, G. S. et al. Waterborne Exophiala species causing disease in cold-blooded animals. Persoonia. 27, 46–72. https://doi.org/10.3767/003158511X614258 (2011).
Zeng, J. S. et al. Spectrum of clinically relevant Exophiala species in the United States. J. Clin. Microbiol. 45, 3713–3720. https://doi.org/10.1128/JCM.02012-06 (2007).
Saraiva, M. et al. Exophiala angulospora infection in hatchery-reared lumpfish (Cyclopterus lumpus) broodstock. J. Fish. Dis. 42, 335–343. https://doi.org/10.1111/jfd.12940 (2019).
Boeger, W. A. et al. Histopathology of the mangrove land crab Ucides cordatus (Ocypodidae) affected by lethargic crab disease. Dis. Aquat. Organ. 78, 73–81. https://doi.org/10.3354/dao01847 (2007).
Biedunkiewicz, A. & Schulz, L. Fungi of the genus Exophiala in tap water—Potential etiological factors of phaeohyphomycoses. Mikologia Lekarska. 19, 23–26 (2012).
Seyedmousavi, S. et al. Exophiala sideris, a novel black yeast isolated from environments polluted with toxic alkyl benzenes and arsenic. Fungal Biol. 115, 1030–1037. https://doi.org/10.1016/j.funbio.2011.06.004 (2011).
Tintelnot, K. et al. Systemic mycosis caused by a new Cladophialophora species. J. Med. Vet. Mycol. 33, 349–354 (1995).
Badali, H. et al. Biodiversity of the genus Cladophialophora. Stud. Mycol. 61, 175–191. https://doi.org/10.3114/sim.2008.61.18 (2008).
Prenafeta-Boldú, F. X. et al. Isolation and characterisation of fungi growing on volatile aromatic hydrocarbons as their sole carbon and energy source. Mycol. Res. 105, 477–484. https://doi.org/10.1017/S0953756201003719 (2001).
Silva, U. C. et al. Long-term rock phosphate fertilization impacts the microbial communities of maize rhizosphere. Front. Microbiol. 8, 1–11. https://doi.org/10.3389/fmicb.2017.01266 (2017).
Xu, J. et al. The structure and function of the global citrus rhizosphere microbiome. Nat. Commun. 9, 4894. https://doi.org/10.1038/s41467-018-07343-2 (2018).
Gomes, E. A. et al. Root-associated microbiome of maize genotypes with contrasting phosphorus use efficiency. Phytobiomes J. 2, 129–137. https://doi.org/10.1094/PBIOMES-03-18-0012-R (2018).
Duarte, A. P. M. et al. Prevalence of the genus Cladosporium on the integument of leaf-cutting ants characterized by 454 pyrosequencing. Antonie Van Leeuwenhoek 109, 1235–1243. https://doi.org/10.1007/s10482-016-0724-3 (2016).
Gomes, S. I. F., Merckx, V. S. F. T. & Saavedra, S. Fungal-host diversity among mycoheterotrophic plants increases proportionally to their fungal-host overlap. Ecol. Evol. 7, 3623–3630. https://doi.org/10.1002/ece3.2974 (2017).
Morais, D., Pylro, V., Clark, I. M., Hirsch, P. R. & Tótola, M. R. Responses of microbial community from tropical pristine coastal soil to crude oil contamination. Peer. J. 18, 1–21. https://doi.org/10.7717/peerj.1733 (2016).
Acknowledgements
This work was supported in CAPES (Coordination for the Improvement of Higher Education Personnel) and CNPq (National Council for Scientific and Technological Development) by grant 400011/2016-6 and Vania Aparecida Vicente received fellowships from CNPq (grant number 312811/2018-7).
Author information
Authors and Affiliations
Contributions
F.F.C. performed the data mining and wrote the manuscript; N.M.S. performed the analysis; M.F.V. wrote the manuscript and helped review; V.A.W. performed the analysis and helped review; L.F.M., G.X.S., R.R.G., R.T.R., M.A.A.C., M.J.N., J.S. and G.B.I.M. helped review; V.A.V. and G.S.H. helped review and modified the manuscript. All authors have reviewed and approved the final content.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Costa, F.d., da Silva, N.M., Voidaleski, M.F. et al. Environmental prospecting of black yeast-like agents of human disease using culture-independent methodology. Sci Rep 10, 14229 (2020). https://doi.org/10.1038/s41598-020-70915-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-020-70915-0
This article is cited by
-
Galleria mellonella in vitro model for chromoblastomycosis shows large differences in virulence between isolates
IMA Fungus (2024)
-
Cryptic fungal diversity revealed by DNA metabarcoding in historic wooden structures at Whalers Bay, Deception Island, maritime Antarctic
Brazilian Journal of Microbiology (2023)
-
Sugarcane: an unexpected habitat for black yeasts in Chaetothyriales
IMA Fungus (2023)