Abstract
Investigation of the prevalence and diversity of Bartonella infections in small mammals in the Qaidam Basin, western China, could provide a scientific basis for the control and prevention of Bartonella infections in humans. Accordingly, in this study, small mammals were captured using snap traps in Wulan County and Ge’ermu City, Qaidam Basin, China. Spleen and brain tissues were collected and cultured to isolate Bartonella strains. The suspected positive colonies were detected with polymerase chain reaction amplification and sequencing of gltA, ftsZ, RNA polymerase beta subunit (rpoB) and ribC genes. Among 101 small mammals, 39 were positive for Bartonella, with the infection rate of 38.61%. The infection rate in different tissues (spleens and brains) (χ2 = 0.112, P = 0.738) and gender (χ2 = 1.927, P = 0.165) of small mammals did not have statistical difference, but that in different habitats had statistical difference (χ2 = 10.361, P = 0.016). Through genetic evolution analysis, 40 Bartonella strains were identified (two different Bartonella species were detected in one small mammal), including B. grahamii (30), B. jaculi (3), B. krasnovii (3) and Candidatus B. gerbillinarum (4), which showed rodent-specific characteristics. B. grahamii was the dominant epidemic strain (accounted for 75.0%). Furthermore, phylogenetic analysis showed that B. grahamii in the Qaidam Basin, might be close to the strains isolated from Japan and China. Overall, we observed a high prevalence of Bartonella infection in small mammals in the Qaidam Basin. B. grahamii may cause human disease, and the pathogenicity of the others Bartonella species needs further study, the corresponding prevention and control measures should be taken into consideration.
Similar content being viewed by others
Introduction
Bartonella is a genus within the Bartonellaceae family in the Alphaproteobacteria class. Bartonella species are small, intracellular, vector-borne hemotropic gram-negative bacteria, some of which can infect a variety of mammals and cause human Bartonellosis1. The Bartonella species and their respective reservoir hosts are increasing constantly, and over 40 species and subspecies of Bartonella have now been detected in domestic and wild animals including cats, dogs, rodents, cattle, deer, bats, and so on2. Several Bartonella species were recognized as human pathogens, such as B. bacilliformis3, B. quintana4, B. henselae5, B. elizabethae6, B. clarridgeiae7, B. koehlerae8, B. vinsonii subsp. arupensis9, B. vinsonii subsp. berkhoffii10, B. grahamii11,12, B. rochalimae13, B. tamiae14, B. ancashensis15, B. washoensis16, and so on, which could cause endocarditis, myocarditis, neuroretinitis, meningitis, splenomegaly, lymphadenopathy and neurologic disorders in humans.
Small mammals, particularly rodents are considered natural reservoirs of many Bartonella species. Previous studies have reported that the infection rate of Bartonella is as high as 70% in rodents worldwide17, including in America18, Asia19, Africa20, and Europe21. Furthermore, many pathogenic Bartonella can be detected in rodents, which indicated that investigating the epidemiological characteristics of Bartonella in rodents is of great significance for the prevention and control of human Bartonellosis.
In the previous study, our team had detected Bartonella species in Ochotona curzoiae in the Qinghai-Tibet Plateau, with a positive rate of 18.99%22. However, few systematic investigations of Bartonella species in small mammals have been reported in the Qinghai-Tibet Plateau. The Qaidam Basin, located in the Haixi Mongolian Tibetan autonomous prefecture, northwest of Qinghai Province and northeastern Qinghai-Tibet Plateau, is the highest basin in China, with an altitude between 2600 and 3000 m; nearly 20 species of rodents have been reported to inhabit this area23. Furthermore, the Qaidam Basin, is a famous tourist attraction, attracting many visitors each year; therefore, the prevention and control of rodent-related pathogenic microorganisms are important for public health.
Accordingly, in this study, we investigated the prevalence and genetic diversity of Bartonella species in small mammals (including rodents and Ochotona curzoiae) from the Qaidam Basin. Our findings provided insights into the distribution of Bartonella in small mammals and the resulting public health threat in this region.
Results
Animal collection
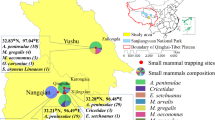
In total, 101 small mammals were captured from six trapping sites, including 78 in Wulan County (01-78 QHWL) and 23 in Ge’ermu City (79-101 QHGEM). All captured small mammals were identified morphologically into eight species, including Cricetulus longicaudatus (46), Mus musculus (17), Phodopus roborovskii (11), Meriones meridians (11), Ochotona curzoniae (10), Allactaga sibirica (3), Cricetulus migratorius (2) and Microtus oeconomus (1). The geographical distribution of the trapped small mammals was shown in Fig. 1.
Bartonella infections
There were 97 spleens and 74 brains of 101 small mammals (excluded uncollected and contaminated specimens) used for Bartonella isolation, and the pure colonies obtained were then confirmed by polymerase chain reaction (PCR) amplification of partial gltA gene (379 bp). In total, 39 small mammals, classified into four species (Cricetulus longicaudatus [29/46], Microtus oeconomus [1/1], Allactaga sibirica [3/3], Meriones meridianus [6/11]), were positive for Bartonella infection; 21 were positive in both the brain and spleen, 12 were positive in the spleen, and six were positive in the brain, with an overall infection rate of 38.61% (39/101). The positive rates in different tissues were not statistically significant (34.02% versus 36.49% for brain and spleen, respectively, χ2 = 0.112, P = 0.738) (Table 1). And the positive rate was 44.64% (25/56) in female and 31.11% (14/45) in male, which the difference was not statistically significant too (χ2 = 1.927, P = 0.165).
There were 45 small mammals of three species captured in farmlands, with a Bartonella infection rate of 40.00% (18/45). Twenty small mammals of three species were captured in forests, with an infection rate of 60.00% (12/20). Additionally, 10 small mammals of one specie were captured in meadows, with no Bartonella infection, and 26 small mammals of four species were captured in semi-desert areas, with an infection rate of 34.62% (9/26). Thus, the infections rates in different habitats were significantly different (χ2 = 10.361, P = 0.016) (Table 2).
Identification of Bartonella species
Through BLAST analysis of the gltA gene, 40 Bartonella strains were obtained, including 18 strains isolated in spleens or brains of different small mammals, 20 strains isolated both in the spleens and brains of the same small mammals, and two strains isolated in the spleen and brain from one small mammal. Overall, 29 isolates from Cricetulus longicaudatus and one isolate from Microtus oeconomus were B. grahamii with 96.75–99.15% identity; three isolates from Allactaga sibirica were B. jaculi with 97.34–97.63% identity; four isolates from Meriones meridianus were Candidatus B. gerbillinarum with 92.06–96.06% identity; and three isolates from Meriones meridianus were B. krasnovii with 97.73–98.57% identity.
We used the maximum likelihood (ML), neighbor-joining (NJ), minimum-evolution (ME), and unweighted pair-group method with arithmetic mean to construct phylogenetic trees and obtained the same results; thus, the ML method was used for further analyses. Phylogenetic trees were constructed based on the DNA sequences of the concatenations of gltA, ftsZ, rpoB and ribC genes (2483 bp), and all isolates clustered into four clusters, i.e., clusters I to IV (Fig. 2). Strains belonging to cluster I were closely related to B. grahamii, strains belonging to cluster II were closely related to B. krasnovii, strains belonging to cluster III were closely related to Candidatus B. gerbillinarum and strains belonging to cluster IV were closely related to B. jaculi. In cluster III, MM82QHGEM was separated from the other three isolates by a long distance and showed 92.06% identity with reference Candidatus B. gerbillinarum, indicating that this isolate might be a new species of Bartonella24.
Overall, our findings indicated that B. grahamii, B. jaculi, B. krasnovi, and Candidatus B. gerbillinarum were prevalent in the Qaidam Basin, and that B. grahamii was the dominant Bartonella species. Bartonella was not detected in the small mammals from two of six trapping sites, and the distribution of Bartonella species showed obvious geographical differences. Moreover, B. grahamii and B. jaculi were distributed in Wulan County, whereas Candidatus B. gerbillinarum and B. krasnovii were distributed in Ge’ermu City (Fig. 3).
Interestingly, our study also showed an association between Bartonella species and small mammal species. B. grahamii was specific for Cricetulus longicaudatus, B. jaculi was specific for Allactaga sibirica, and Candidatus B. gerbillinarum and B. krasnovii were specific for Meriones meridianus. In addition, two Bartonella species, i.e., Candidatus B. gerbillinarum and B. krasnovii were isolated from different tissues in one Meriones meridianu. Candidatus B. gerbillinarum was isolated from the brain, and B. krasnovii was isolated from the spleen, suggesting that one rodent could carry more than one Bartonella species (Table 3). It indicated the coinfection phenomenon existed in Bartonella species, which was consistent with previous studies25,26.
Phylogenetic analysis
Phylogenetic analysis based on gltA sequences showed that B. grahamii in the Qaidam Basin was mainly clustered into three clusters. Some strains from Cricetulus longicaudatus clustered with B. grahamii from Apodemus speciosus in Japan, some strains from Cricetulus longicaudatus clustered with B. grahamii from Ochotona curzoniae in China, and one strain from Microtus oeconomus was clustered separately, indicating the genetic diversity of B. grahamii prevalent in the Qaidam Basin.
Three strains of AS61QHWL-AS63QHWL from Allactaga sibirica were clustered with B. jaculi from Jaculus orientails in Egypt, four strains of MM79BQHGEM, MM82QHGEM, MM84QHGEM and MM85QHGEM from Meriones meridianus were clustered with Candidatus B. gerbillinarum from Synosternus cleopatrae, Gerbillus andersoni and Gerbillus pyramidum in Israel, and three strains of MM79SQHGEM, MM80QHGEM, and MM83QHGEM from Meriones meridianus were clustered with B. krasnovii from Synosternus cleopatrae, Gerbillus andersoni and Gerbillus pyramidum in Israel. The results showed that the Qaidam Basin strains of B. jaculi, Candidatus B. gerbillinarum and B. krasnovii were clustered with the relative reference strains, but formed distinct branches, except MM80QHGEM. Thus, these findings suggested that Bartonella infection was rodent specific and continued to evolve (Fig. 4).
Discussion
Bartonella species are highly prevalent in small mammals worldwide, which have the close contact with humans. It is of great significance to investigate the epidemiological and ecological characteristics of Bartonella infection in small mammals from different areas. In this study, we observed the prevalence and genetic diversity of Bartonella species in small mammals in the Qaidam Basin. The infection rate of Bartonella species in small mammals was 38.61%, which was higher than that of 18.99% in Ochotona curzoniae in our previous study22, and higher than that in most areas of China27, but lower than that in many other countries, including Russia (60–83%), Canada (48–90%), Netherlands (72%), etc17. In addition, the infection rate was significantly different in different habitats (farmlands, forests, meadows, and semi-desert areas), with the highest infection rates observed in forests.
The nutritional requirements of Bartonella make it difficult to culture in vitro28. Generally, the spleen tissue is used in Bartonella culture. In this study, brain tissue was successfully used for Bartonella isolation for the first time. Additionally, we found that the positive rates in different tissues (spleens and brains) of small mammals did not differ significantly. Moreover, we detected two Bartonella species in the same small mammal, i.e., B. krasnovii in the spleen and Candidatus B. gerbillinarum in the brain, indicating the complexity of Bartonella infection in small mammals. A previous study indicated that there might be a high Bartonella coinfection rate in rodents29. Therefore, in order to further explore the coinfection of Bartonella species, additional studies of multi-tissue culture, multi-clone detection, and multiple PCR detection by using well-defined species or genotype PCR primer sets are needed25,26,29.
Several rodent-associated Bartonella species have been implicated as the causative agents of human disease. Here, we obtained four Bartonella species in rodents, B. grahamii, B. jaculi, B. krasnovii and Candidatus B. gerbillinarum. Especially, B. grahamii was detected in Cricetulus longicaudatus specificly, B. jaculi was detected in Allactaga sibirica specificly, Candidatus B. gerbillinarum and B. krasnovii were detected in Meriones meridianus specificly, indicating rodent-specific characteristics. B. grahamii was the dominant epidemic strain circulating in the Qaidam Basin, which was associated with neuroretinitis and cat scratch disease (CSD) in immunocompromised people11,12, suggesting that Bartonella species detected in Cricetulus longicaudatus may have the ability to cause human disease. It was reported that Bartonella species antibodies and DNA were detected in cerebrospinal fluid of cats and dogs30,31, suggesting the possible relationship between Bartonella infection and central nervous system disease. Bartonella isolation from the rodent brain supported this observation. Until now, B. henselae and B. quintana were reported to cause central nervous system infection32,33, however, the effects of B. grahamii on the central nervous system needs further investigation.
Through phylogenetic analysis based on gltA sequences collected from the GenBank, B. grahamii from Cricetulus longicaudatus in the Qaidam Basin was mainly clustered with B. grahamii from Apodemus speciosus in Japan34 and Ochotona curzoniae in China22. Some B. grahamii prevalent in the Qaidam Basin might have high homology with strains from Japan, which was consistent with the previous study34. B. jaculi, Candidatus B. gerbillinarum and B. krasnovii have only been reported in Eygpt35 and Israel29, which pathogenicity was not clear and needs further study. However, in this study, Bartonella infection was not detected in Ochotona curzoniae, possibly because of the differences in sampling sites and the small number of samples collected, which need further investigation.
Conclusions
This study provided a better understanding of the prevalence and genetic diversity of Bartonella species in small mammals from the Qaidam Basin. Four Bartonella species were detected in rodents, among which B. grahamii was the dominant strain, and potentially pathogenic to humans. Additionally, Bartonella were isolated from rodent brains for the first time, and two Bartonella species were detected in different tissues of the same rodent, which indicated the complexity of Bartonella infection and the necessity for multi-tissue culture, multi-clone detection and multiple PCR detection. Our results raise the potential threats to public health by the Bartonella species, and surveillance of Bartonella in animals and investigation of suspected clinical cases in humans need to be strengthened in the Qaidam Basin.
Materials and methods
Ethical statement
This study was approved by the Ethics Committee of Chinese Center for Disease Control and Prevention (No: ICDC-2015001). Small mammals were sampled with the help of Wulan Center for Disease Control and Prevention, and Ge’ermu Center for Disease Control and Prevention. All animals were treated according to the Guidelines of Regulations for the Administration of Laboratory Animals (Decree No. 2 of the State Science and Technology Commission of the People's Republic of China, 1988) and the Guidelines for Treating Animals Kindly from Ministry of Science and Technology of the People’s Republic of China. All efforts were made to minimize discomfort to the animals.
Sample collection
According to the previous study36, small mammals were captured using snap traps in a diversity of habitats in July 2018 in Wulan County (latitude 36.32°–37.33°N, longitude 97.02°–99.45°E) and Ge’ermu City (latitude 35.18°–37.80°N, longitude 91.72°–95.85°E), Qinghai Province. And the small mammal species were determined morphologically. The spleens and brains were harvested under sterile conditions from each animal after euthanasia, and all samples were stored in liquid nitrogen until use.
Bartonella culture
Approximately 20 mg of each spleen and brain sample from the small mammal was homogenized by adding 200 μL sterilized trypsin soy broth (BD Biosciences, Franklin Lakes, NJ, USA), plated onto two trypsin soy agars containing 5% (vol/vol) defiber sheep blood (BD Biosciences), and incubated at 37 °C in an atmosphere containing 5% CO2. Pure colonies of Bartonella species were obtained according to previous methods22.
DNA extraction, PCR amplification and DNA sequencing
Crude DNA was extracted according to the previous method22. PCR was performed to detect the Bartonella citrate synthase (gltA) gene. Then, gltA positive strains were evaluated for amplification of ftsZ, RNA polymerase beta subunit (rpoB) and ribC genes. DNA amplification was performed in 50 μL mixtures containing 25 μL 2 × TransTaq-T PCR SuperMix (Beijing TransGen Biotech Co., Ltd., Beijing, China), 22 μL double-distilled H2O, 1 μL (10 μmol/L) of each primer (sequences listed in Table 4), and 1 μL of DNA template. gltA amplification was performed under the following conditions: one cycle for 5 min at 94 °C; 33 cycles for 30 s at 94 °C, 30 s at 53 °C, and 20 s at 72 °C; and a final extension for 7 min at 72 °C. The annealing temperatures for amplification of the ftsZ, rpoB and ribC were 55 °C, 50 °C and 50 °C, respectively. Next, 5 μL of each PCR product was run on 1% agarose gels, stained with ethidium bromide, and visualized using a gel imaging system (Bio-Rad, Hercules, CA, USA). The expected PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols, and then sequenced using specific primers for gltA, ftsZ, rpoB and ribC with an Applied Biosystems 3730 xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Phylogenetic analysis
The sequences generated in this study were submitted to the GenBank (accession numbers MT815283-MT815438). The nucleotide sequence homology was blasted against reported Bartonella species sequences in the GenBank using the BLAST program at the National Center for Biotechnology Information Website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic tree was created using the maximum-likelihood method with MEGA version 7.0, and bootstrap values were calculated with 1000 replicates41,42 (incomplete and poor quality sequences were excluded from phylogenetic analysis). Brucella abortus was used as the outgroup.
Statistical analysis
The positive rates of Bartonella in different habitats, genders and tissues of small mammals were analyzed using the Chi-square test. All data were analyzed using SPSS 22.0 (SPSS, Inc., Chicago, IL, USA). P < 0.05 was considered statistically significant.
Consent to publish
All the authors consent to publish the article in its present form.
Data availability
The data supporting the conclusions of this article are included within the article.
References
Mogollon-Pasapera, E., Otvos, L. Jr., Giordano, A. & Cassone, M. Bartonella: emerging pathogen or emerging awareness?. Int. J. Infect. Dis. 13, 3–8. https://doi.org/10.1016/j.ijid.2008.04.002 (2009).
Huang, K. et al. Molecular detection of Bartonella spp. in China and St. Kitts. Can. J. Infect. Dis. Med. Microbiol. 2019, 3209013. https://doi.org/10.1155/2019/3209013 (2019).
Minnick, M. F. et al. Oroya fever and verruga peruana: bartonelloses unique to South America. PLoS Negl. Trop. Dis. 8, e2919. https://doi.org/10.1371/journal.pntd.0002919 (2014).
Arvand, M., Raoult, D. & Feil, E. J. Multi-locus sequence typing of a geographically and temporally diverse sample of the highly clonal human pathogen Bartonella quintana. PLoS ONE 5, e9765. https://doi.org/10.1371/journal.pone.0009765 (2010).
Chaudhry, R. et al. Bartonella henselae infection in diverse clinical conditions in a tertiary care hospital in north India. Indian J. Med. Res. 147, 189–194. https://doi.org/10.4103/ijmr.IJMR_1932_16 (2018).
Daly, J. S. et al. Rochalimaea elizabethae sp. Nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 31, 872–881. https://doi.org/10.1128/JCM.31.4.872-881.1993 (1993).
Kordick, D. L. et al. Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch disease). J. Clin. Microbiol. 35, 1813–1818. https://doi.org/10.1128/JCM.35.7.1813-1818.1997 (1997).
Avidor, B. et al. Bartonella koehlerae, a new cat-associated agent of culture-negative human endocarditis. J. Clin. Microbiol. 42, 3462–3468. https://doi.org/10.1128/JCM.42.8.3462-3468.2004 (2004).
Fenollar, F., Sire, S. & Raoult, D. Bartonella vinsonii subsp. arupensis as an agent of blood culture-negative endocarditis in a human. J. Clin. Microbiol. 43, 945–947. https://doi.org/10.1128/JCM.43.2.945-947.2005 (2005).
Breitschwerdt, E. B. et al. Bartonella vinsonii subsp. berkhoffii and Bartonella henselae bacteremia in a father and daughter with neurological disease. Parasit. Vectors 3, 29. https://doi.org/10.1186/1756-3305-3-29 (2010).
Kerkhoff, F. T., Bergmans, A. M., van Der Zee, A. & Rothova, A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J. Clin. Microbiol. 37, 4034–4038. https://doi.org/10.1128/JCM.37.12.4034-4038.1999 (1999).
Oksi, J. et al. Cat scratch disease caused by Bartonella grahamii in an immunocompromised patient. J. Clin. Microbiol. 51, 2781–2784. https://doi.org/10.1128/JCM.00910-13 (2013).
Chomel, B. B. et al. Dogs are more permissive than cats or guinea pigs to experimental infection with a human isolate of Bartonella rochalimae. Vet. Res. 40, 27. https://doi.org/10.1051/vetres/2009010 (2009).
Colton, L., Zeidner, N., Lynch, T. & Kosoy, M. Y. Human isolates of Bartonella tamiae induce pathology in experimentally inoculated immunocompetent mice. BMC Infect. Dis. 10, 229. https://doi.org/10.1186/1471-2334-10-229 (2010).
Mullins, K. E. et al. Whole-genome analysis of bartonella ancashensis, a novel pathogen causing Verruga Peruana, Rural Ancash Region, Peru. Emerg. Infect. Dis. 23, 430–438. https://doi.org/10.3201/eid2303.161476 (2017).
Kosoy, M., Murray, M., Gilmore, R. D. Jr., Bai, Y. & Gage, K. L. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J. Clin. Microbiol. 41, 645–650. https://doi.org/10.1128/jcm.41.2.645-650.2003 (2003).
Gutierrez, R. et al. Bartonella infection in rodents and their flea ectoparasites: an overview. Vector Borne Zoonotic Dis. 15, 27–39. https://doi.org/10.1089/vbz.2014.1606 (2015).
Kosoy, M. Y. et al. Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United States. Am. J. Trop. Med. Hyg. 57, 578–588. https://doi.org/10.4269/ajtmh.1997.57.578 (1997).
Bordes, F. et al. The diversity of microparasites of rodents: a comparative analysis that helps in identifying rodent-borne rich habitats in Southeast Asia. Infect. Ecol. Epidemiol. https://doi.org/10.3402/iee.v3i0.20178 (2013).
Pretorius, A. M., Beati, L. & Birtles, R. J. Diversity of bartonellae associated with small mammals inhabiting Free State province, South Africa. Int. J. Syst. Evol. Microbiol. 54, 1959–1967. https://doi.org/10.1099/ijs.0.03033-0 (2004).
Gundi, V. A. et al. Isolation of Bartonella rattimassiliensis sp nov and Bartonella phoceensis sp. Nov. from European Rattus norvegicus. J. Clin. Microbiol. 42, 3816–3818. https://doi.org/10.1128/JCM.42.8.3816-3818.2004 (2004).
Rao, H. X. et al. Bartonella species detected in the plateau pikas (Ochotona curzoiae) from Qinghai Plateau in China. Biomed. Environ. Sci. 28, 674–678. https://doi.org/10.3967/bes2015.094 (2015).
Li, H. L. et al. Investigation of geographical distribution pattern of rodents in Qinghai province, China. Chin. J. Vector Biol. Control 24, 418–421 (2013).
Blazes, D. L. et al. Novel Bartonella agent as cause of verruga peruana. Emerg. Infect. Dis. 19, 1111–1114. https://doi.org/10.3201/eid1907.121718 (2013).
Breitschwerdt, E. B. & Maggi, R. G. Bartonella quintana and Bartonella vinsonii subsp. vinsonii bloodstream co-infection in a girl from North Carolina, USA. Med. Microbiol. Immunol. 208, 101–107. https://doi.org/10.1007/s00430-018-0563-0 (2019).
Diniz, P. P. et al. Canine bartonellosis: serological and molecular prevalence in Brazil and evidence of co-infection with Bartonella henselae and Bartonella vinsonii subsp. berkhoffii. Vet. Res. 38, 697–710. https://doi.org/10.1051/vetres:2007023 (2007).
Liu, Q., Eremeeva, M. E. & Li, D. Bartonella and Bartonella infections in China: from the clinic to the laboratory. Comp. Immunol. Microbiol. Infect. Dis. 35, 93–102. https://doi.org/10.1016/j.cimid.2012.01.002 (2012).
Li, D. M., Miao, Z. G., Song, X. P., Wang, J. & Liu, Q. Y. Optimization of liquid growth conditions and determination of growth curves for Bartonella species. Microbiol. China 39, 1695–1702 (2012).
Gutierrez, R. et al. Untangling the knots: co-infection and diversity of Bartonella from wild gerbils and their associated fleas. Mol. Ecol. 27, 4787–4807. https://doi.org/10.1111/mec.14906 (2018).
Leibovitz, K., Pearce, L., Brewer, M. & Lappin, M. R. Bartonella species antibodies and DNA in cerebral spinal fluid of cats with central nervous system disease. J. Feline Med. Surg. 10, 332–337. https://doi.org/10.1016/j.jfms.2007.12.011 (2008).
Bartner, L. R. et al. Testing for Bartonella ssp. DNA in cerebrospinal fluid of dogs with inflammatory central nervous system disease. J. Vet. Intern. Med. 32, 1983–1988. https://doi.org/10.1111/jvim.15288 (2018).
Podsiadly, E., Chmielewski, T. & Tylewska-Wierzbanowska, S. Bartonella henselae and Borrelia burgdorferi infections of the central nervous system. Ann. N. Y. Acad. Sci. 990, 404–406. https://doi.org/10.1111/j.1749-6632.2003.tb07400.x (2003).
Parrott, J. H. et al. Central nervous system infection associated with Bartonella quintana: a report of two cases. Pediatrics 100, 403–408. https://doi.org/10.1542/peds.100.3.403 (1997).
Inoue, K. et al. Evolutional and geographical relationships of Bartonella grahamii isolates from wild rodents by multi-locus sequencing analysis. Microb. Ecol. 57, 534–541. https://doi.org/10.1007/s00248-009-9488-x (2009).
Sato, S. et al. Bartonella jaculi sp. nov., Bartonella callosciuri sp. nov., Bartonella pachyuromydis sp. nov. and Bartonella acomydis sp. nov., isolated from wild Rodentia. Int. J. Syst. Evol. Microbiol. 63, 1734–1740. https://doi.org/10.1099/ijs.0.041939-0 (2013).
Qin, X. R., Liu, J. W., Yu, H. & Yu, X. J. Bartonella species detected in rodents from Eastern China. Vector Borne Zoonotic Dis. 19, 810–814. https://doi.org/10.1089/vbz.2018.2410 (2019).
Norman, A. F., Regnery, R., Jameson, P., Greene, C. & Krause, D. C. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 33, 1797–1803. https://doi.org/10.1128/JCM.33.7.1797-1803.1995 (1995).
Zeaiter, Z., Liang, Z. & Raoult, D. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsZ gene sequences. J. Clin. Microbiol. 40, 3641–3647. https://doi.org/10.1128/jcm.40.10.3641-3647.2002 (2002).
Renesto, P., Gouvernet, J., Drancourt, M., Roux, V. & Raoult, D. Use of rpoB gene analysis for detection and identification of Bartonella species. J. Clin. Microbiol. 39, 430–437. https://doi.org/10.1128/JCM.39.2.430-437.2001 (2001).
Johnson, G., Ayers, M., McClure, S. C., Richardson, S. E. & Tellier, R. Detection and identification of Bartonella species pathogenic for humans by PCR amplification targeting the riboflavin synthase gene (ribC). J. Clin. Microbiol. 41, 1069–1072. https://doi.org/10.1128/jcm.41.3.1069-1072.2003 (2003).
Bai, Y. et al. Global distribution of Bartonella infections in domestic bovine and characterization of Bartonella bovis strains using multi-locus sequence typing. PLoS ONE 8, e80894. https://doi.org/10.1371/journal.pone.0080894 (2013).
Huang, R. et al. Bartonella quintana infections in captive monkeys, China. Emerg. Infect. Dis. 17, 1707–1709. https://doi.org/10.3201/eid1709.110133 (2011).
Acknowledgements
We wish to express our gratitude to Wulan County Center for Disease Control and Prevention and Ge’ermu City Center for Disease Control and Prevention employees, who participated in the specimen collection.
Funding
The present work was supported by Natural Science Foundation of Shanxi Province of China (201901D111326), Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (2020L0376, 2020L0377), Major Infectious Diseases Such as AIDS and Viral Hepatitis Prevention and Control Technology Major Projects (2018ZX10712001, 2017ZX10303404), Doctor research startup foundation of Changzhi Medical College (BS201912, BS201921), Guiding Project of Qinghai Health and Family Planning Commission (2018-wjzdx-82) and Qinghai Thousand People Plan.
Author information
Authors and Affiliations
Contributions
D.M.L. and J.Y. conceived and designed the experiments. D.M.L. and X.P.S. performed the experiments. H.X.R., J.Y. and D.M.L. analyzed the data. S.J.L., L.L., R.W., K.S. and Y.S. contributed the samples. L.L. identified the small mammals. H.X.R., J.Y. and D.M.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rao, H., Li, S., Lu, L. et al. Genetic diversity of Bartonella species in small mammals in the Qaidam Basin, western China. Sci Rep 11, 1735 (2021). https://doi.org/10.1038/s41598-021-81508-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-021-81508-w
This article is cited by
-
Genetic diversity of Bartonella species in small mammals in the Qinghai Menyuan section of Qilian Mountain National Park, China
Scientific Reports (2024)
-
Emerging rodent-associated Bartonella: a threat for human health?
Parasites & Vectors (2022)
-
Detection and genetic diversity of Bartonella species in small mammals from the central region of the Qinghai-Tibetan Plateau, China
Scientific Reports (2022)