Abstract
In cytological examination, suspicious cells are evaluated regarding malignancy and cancer type. To assist this, we previously proposed an automated method based on supervised learning that classifies cells in lung cytological images as benign or malignant. However, it is often difficult to label all cells. In this study, we developed a weakly supervised method for the classification of benign and malignant lung cells in cytological images using attention-based deep multiple instance learning (AD MIL). Images of lung cytological specimens were divided into small patch images and stored in bags. Each bag was then labeled as benign or malignant, and classification was conducted using AD MIL. The distribution of attention weights was also calculated as a color map to confirm the presence of malignant cells in the image. AD MIL using the AlexNet-like convolutional neural network model showed the best classification performance, with an accuracy of 0.916, which was better than that of supervised learning. In addition, an attention map of the entire image based on the attention weight allowed AD MIL to focus on most malignant cells. Our weakly supervised method automatically classifies cytological images with acceptable accuracy based on supervised learning without complex annotations.
Similar content being viewed by others
Introduction
Lung cancer is the leading cause of cancer death among males and females worldwide, and early and accurate diagnosis is critical for survival1. When the presence of a tumor is noted in a chest X-ray or CT examination, pathological examination is performed to classify it as benign or malignant2. Pathological examination includes cytology and histology. In cytology, a cytotechnologist (screener) first screens the samples and finds suspicious cells. The cytopathologist then observes the identified cells in detail and a decision is sometimes made using a combination of regular cytology specimens and immunostaining specimens. Cytology, the first step in detecting malignant cells, plays an important role in influencing the final diagnostic results. However, the screener must find suspicious cells within a large number of cells. The task is extremely burdensome, and there are concerns about the variation in diagnostic accuracy among screeners. Therefore, in this study, we focused on the classification of benign and malignant lung cells based on deep learning technology, which has excellent image recognition capability.
Deep learning technology is an application of multilayer neural network technology and convolutional neural networks (CNNs), which were developed with inspiration from the workings of vision, and is widely used for image classification, object detection, and prediction. There are also many applications in medical imaging3,4,5,6,7,8,9. For pathological images, CNN has also been used to determine the distribution of malignant cells10,11 and to classify cells12,13. We have developed diagnostic support technology for lung cytology, and have proposed methods that use CNN to classify cells as benign or malignant14.
In these studies, target image and label (correct answer) pairs for that image are given to the deep learning model for training, which is called supervised learning. Good classification performance is achieved when the correct label is assigned to each image. On the contrary, if the labels are inaccurate, the classification performance will be degraded. In cytology, there are some similarities between benign and malignant cells, and there are also many atypical cells. Therefore, it is difficult to assign accurate labels of benign and malignant to individual cells. Recently, multiple instance learning (MIL) has been attracting attention as a weakly supervised learning method that can train networks without creating labels on a one-to-one basis15.
MIL introduces the concept of “instances” that represent individual data and “bags” that contain instances that belong to groups such as cases. The network performs the classification in bag units. In this case, even if there are extra or incorrect instances in the bag that do not contribute to the classification, their influence is low because the classification is performed comprehensively on a bag-by-bag basis. Recently, several methods that introduce attention mechanisms and deep learning16 have been proposed17,18. An attention mechanism is a technique to evaluate which information should be paid attention to when a target contains a large amount of information. Recently, it has been widely used in the fields of natural language processing and image recognition. By clarifying the areas to be focused on, better performance can be obtained compared to that obtained using conventional methods. By visualizing the internal parameters of the attention mechanism, it is possible to determine the instances that have been paid attention to, and to visualize the basis for the decision. Deep learning techniques are also effective in extracting features from instances and processing attention mechanisms. Ilse et al. proposed an attention-based deep MIL (AD MIL) using deep learning and the attention mechanism, and showed that it can be applied to text and cell classifications in histological images18. In addition, Hashimoto et al. applied MIL processing of whole slide images at multiple scales to classify lymphoma subtypes17.
To the best of our knowledge, there are no studies on the classification of benign and malignant cells in lung cytological images using weakly supervised learning, and no comparison has been made with supervised learning. In this study, we developed a method for the classification of benign and malignant cytological images using AD MIL and compared its performance with that of supervised learning.
Methods
Outline of proposed scheme
An outline of the proposed method is shown in Fig. 1. Images of lung cytological specimens were divided into small patch images and stored in bags with case IDs. Each bag was then labeled as benign or malignant, and the classification of benign or malignant was performed by supplying the bag units to the AD MIL.
Image dataset
This study was performed as a retrospective study with permission from the Institutional Review Board of Fujita Health University (IRB number: HM16-155). The informed consent was obtained from patients subject to the condition of data anonymization. All experimental protocols were performed in accordance with the relevant guidelines and regulations in compliance with the Declaration of Helsinki. For this study, lung cells of 322 patients were collected with interventional cytology using either bronchoscopy or CT-guided fine-needle aspiration cytology, and comprised 108 benign and 214 malignant cases. Malignant cases comprised 124 adenocarcinomas, 52 squamous cell carcinomas, and 38 small cell carcinomas. In these diagnoses, a final decision was made in conjunction with the histological analysis of specimen from a biopsy. Biopsy tissues were collected simultaneously with cytological specimens, fixed in 10% formalin, dehydrated, and embedded in paraffin. In some cases, where diagnosis was difficult, the 3 μm tissue sections were subjected to immunohistochemical analysis to make the decision. Cytological specimens were prepared with liquid-based cytology using the BD SurePathTM liquid-based Pap test (Beckton Dickinson, Franklin Lakes, NJ, USA) and were stained using the Papanicolaou method. Using a digital camera (DP20, Olympus Corporation, Tokyo, Japan) attached to a microscope (BX53, Olympus Corporation) with a 40 × objective lens, 1252 microscopic images of benign cells and 1805 of malignant cells were acquired in a JPEG format with a size of 1280 × 960 pixels per image.
To classify these images, it is important to understand the characteristics of the cells of interest. The characteristics of benign and malignant cells are described below.
Characteristics of benign cells
Benign cells have smaller, more uniform nuclei, and smoother cell borders. Nucleoli are small and few. The volume of cytoplasm is large, and normal pulmonary bronchial epithelial cells have cilia.
Characteristics of malignant cells
The characteristics of malignant cells vary greatly, depending on their tissue type (adenocarcinoma, squamous cell carcinoma, small cell carcinoma). In general, malignant cells have less cytoplasm and irregularly shaped nuclei. In adenocarcinoma, large nucleoli are often observed and the chromatin particles are rough. In addition, nuclei are often unevenly distributed in the cytoplasm. Squamous cell carcinoma, which is often confused with adenocarcinoma, has smaller nucleoli and finer chromatin particles. The cytoplasm is thick, and the nuclei are often located near the center of the cytoplasm. In these two histological types, the nuclei are larger than in normal cells, whereas in small cell carcinoma, the nuclei are smaller and are often interconnected. On Papanicolaou staining, the nuclei of small cell carcinoma cells are darker than those of other histological types and appear different in color from other cells.
Generation of instances
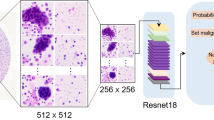
Because MIL can classify a batch of data based on a large amount of data, each specimen image with a matrix size of 1280 × 960 was divided into small patches that were used as instance candidates (Fig. 2). For the matrix size of patch images, we used 64 × 64 pixels as the baseline, as the larger cell nuclei can be included in the image. We also evaluated the classification performance with the matrix sizes of 96 × 96 and 128 × 128 pixels. Here, the image of the captured specimen had a wide field of view, and the patch images contained areas without cells. Therefore, the image was binarized to divide it into regions with cells and background regions. If the area of cells occupied more than 10% of the area in the patch image of the instance candidate, the instance was registered as a malignant or benign bag for analysis; otherwise, it was deleted. The binarization threshold was calculated for each image using the Otsu's automatic binarization algorithm.
Classification method
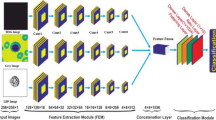
The structure of the AD MIL used for classification is shown in Fig. 318, and consisted of a backbone layer, an MIL attention layer, and one output layer (Fig. 3a). The backbone layer was composed of a CNN, and by providing the instance (Ik) in the bag into the CNN, feature extraction was performed, and the feature vector hk was obtained. Here, we employed LeNet-like19, AlexNet-like20, inception architecture (Inception)21, networks with residual structure (ResNet)22, and dense connected structure (DenseNet)23 (Fig. 3b) as CNN in the backbone layer, and compared their classification performance. The structure of the CNN was determined empirically.
In the conventional MIL technique, the synthesized vector, which is obtained by calculating the maximum or average value of the elements of the feature vector group in the bag, was used for identification. However, the importance of an instance could not be accurately recognized with the simple calculation of the maximum or average values in the conventional method. Therefore, Ilse et al. proposed an MIL technique using an attention mechanism18. Attention mechanism allows the control of the neural network to pay more attention on instances that are most likely to be labeled as positive. It detects key information from a large amount of inaccurate data, which is consistent with the practical diagnosis process.
In this method, a neural network is used to calculate the attention weight, which represents the importance of an instance, and a weighted feature vector z, which is a weighted average of the instances, is obtained using the following equations18.
where hk is the feature vector of the kth instance, and ak the attention weight given for each instance and normalized so that the total value of ak is 1 for each bag. Two vectors of w and V are the parameters for calculating the attention weight, which are determined by training the network. The weighted features z were then assigned to a fully connected (FC) layer with a single artificial neuron, and bag classification (benign and malignant) was performed. Here, a sigmoid function was introduced as the activation function of the artificial neuron. Therefore, AD MIL weights the instances in the bag to create a single vector that is representative of the bag, which is then used to identify whether the bag is benign or malignant. The code of the AD MIL used in this study was a modified version of the code proposed by Ilse et al.18, and we used a Python program developed using Keras and Tensorflow as APIs for deep learning.
Evaluation metrics
To confirm the effectiveness of the proposed method, we evaluated its classification accuracy. All cases were used to evaluate the ability of classification between benign and malignant on a case-by-case basis using the tenfold cross-validation method. Cross-validation is a method that divides data into multiple datasets, and repeatedly trains and evaluates them to obtain classification accuracy for all data. In this study, we divided the data into 10 sets, used 9 of the 10 sets as training data, and evaluated the remaining dataset, which was repeated 10 times. In each training, 10% of the training data were used as validation data while monitoring the training error. The number of training epochs was 100, the batch size was 1, and Adam24 was used as the optimization algorithm. The learning rates, β1, and β2, were 0.0005, 0.9, and 0.999, respectively. We also used a computer with an Intel core i7 7800X CPU and an NVIDIA Quadro RTX 8000 GPU for training.
This method was aimed at improving the performance of MIL by using the attention mechanism. To confirm the effectiveness of the attention mechanism, we compared the classification performance with the attention mechanism disabled. Specifically, as in the conventional MIL, the feature vectors extracted from each instance were averaged, and the combined vector was calculated and used for classification.
Furthermore, to compare our method with supervised learning, we performed general supervised learning on data where all case images contained labels (benign or malignant). We prepared a CNN with an AlexNet-like structure, which was also used in the AD MIL model, with one additional FC layer, and classified it as benign or malignant using the Softmax function. Then, we trained and evaluated the prediction results for each image using the tenfold cross-validation method, as in the evaluation of the AD MIL. Based on the obtained prediction results, we calculated the confusion matrix and classification accuracy by calculating the prediction probability for each image. Note that AD MIL was evaluated on a case-by-case basis, and in actual clinical practice, if even one malignant cell is included in an image, it is considered malignant. Therefore, in the classification by supervised learning, the maximum value of the malignancy probability was obtained for all the images included in one case13. Based on the maximum probability, benign and malignant images were classified, and the overall classification accuracy per case was evaluated. The number of training epochs was 100 and the batch size was 16. We also employed Adam optimization algorithm with the learning rates of 0.00005, β1 of 0.9, and β2 of 0.999. The same parameters as in MIL were used, except for a batch size of 16. Then, for cases classified as malignant by AD MIL, the distribution of attention weights was determined for the instances comprising a single image, and a color attention map was obtained. This allowed us to analyze which part of the specimen was the focus.
Results
Figure 4 shows the accuracy of the proposed method for classifying benign and malignant patches while changing the matrix size of the patch images. As a result, the best performance was obtained when the matrix size of the patch image was set to 64 × 64 pixels, which was used in subsequent analyses.
Tables 1, 2, 3, 4, 5, 6 and 7 show the confusion matrices obtained by summing up the results of classification by AD MIL while changing the CNN structure, and the results of image-based and case-based classification by supervised learning. Sensitivity, specificity, accuracy, and balanced accuracy are shown in Table 8. Here, balanced accuracy is expressed as the average of sensitivity and specificity, and is used when the number of images in the data differs between two classes. In the evaluation results, the classification by AD MIL, which adopts an AlexNet-like structure for CNN, showed the best classification accuracy.
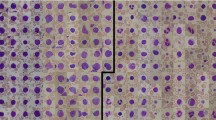
An example of the overall microscopic images of benign cases that were correctly and incorrectly classified by the AD MIL is shown in Fig. 5. In the right side of paired images are attention maps drawn by the attention weights of all instances. Similarly, the image of a malignant case is shown in Fig. 6.
Discussion
In this study, we developed a method for the classification of benign and malignant lung cytological images using AD MIL, a weakly supervised learning method. Three CNN models for the AD MIL were compared, and the AlexNet-like model showed the best classification performance, with an accuracy of 0.916. For general image classification tasks, inception and dense model, which have complex structures, show good performance. These are mainly trained using 224 × 224 pixels as input and a large number of images. In the comparative experiments conducted in this study, the AlexNet-like model with a simple structure showed the best performance. In this study, the weights in the network are trained from the initial state to extract image features from images as small as 64 × 64 pixels. From the experimental results, it is considered that the simpler architecture performed better feature extraction than the more complex one.
Upon comparing the performance of the proposed method, with and without the attention mechanism, the results of the method with the attention mechanism (CNN: AlexNet-like model) showed up to 9% higher classification performance with balanced accuracy, indicating that the attention mechanism is effective for cell classification. The results showed that the attention mechanism was effective in classifying cells.
As a supervised learning method, image classification was performed with the AlexNet-like CNN model used in AD MIL, and the results show that AD MIL was slightly better than supervised learning. In the attention maps presented in Figs. 5 and 6, the focus of attention is slightly different between malignant and benign cases. In malignant cases, the cells that are strongly suspected to be malignant are at a high attention level (yellow or red). On the contrary, in benign cases, attention tended to be paid to the whole area or to the periphery of the cells. Benign cells have a relatively uniform shape of nuclei and may show striae around the cytoplasm; it is believed that AD MIL has been trained to classify the abovementioned differences. Even when a benign image is incorrectly classified as malignant, the cytotechnologist or cytopathologist only needs to confirm the area focused on by AD MIL, which may shorten the diagnosis time.
The best matrix size for the instance (patch image) given to AD MIL was 64 × 64 pixels. This is thought to be the size of the field of view where a single cell fits in the microscopic image and attention to individual cells is engaged.
Both malignant and benign cases contain a variety of cell types. For malignant cells, as mentioned above, the shape of the nucleus and cytoplasm, as well as the structure, arrangement, and color of the inside of the nucleus, differ from those of benign cells. In addition, there are three subcategories of lung cancer, each of which has different characteristics and incidence rates. The breakdown of the types of lung cancer used in this study is 58% adenocarcinoma, 24% squamous cell carcinoma, and 18% small cell carcinoma; these percentages reflect the actual percentage of patients. Because the proposed method could be used for correct classification of benign and malignant cells among these case groups, it appears to have identified the characteristics contained in the three types of malignant cells and discriminated malignant cells from benign cells.
In our previous studies, classification of benign and malignant lung cytological images was performed with nearly 90% accuracy. If experts such as cytolopathologists accurately label individual cytological images as benign or malignant, good classification performance can be achieved. However, a cytological specimen obtained from a single patient contains a large number of cells, and it is extremely difficult to designate the benign and malignant properties of each cell. In addition, when experts evaluate these specimens, they do not classify them as benign or malignant on a cell-by-cell basis, but rather make a comprehensive diagnosis, and thus the classification methods used by experts and supervised learning are different. Rather than assigning a correct label to each cell, our approach was to divide a large number of cell images obtained from a single patient into small patch images (instances) and classify them comprehensively on a bag-by-bag basis. In addition, CNN was used to obtain advanced features from the images, and an attention mechanism was used to select and evaluate the images of interest. Therefore, this method is closer to the clinical diagnosis procedure than conventional supervised learning: it can classify a large number of images with high accuracy without the need for complicated work by experts.
The field of view in this study was relatively small because the images were collected by a camera attached to a microscope; this method may be more useful when using whole slide images containing a large number of cells.
A limitation of this study was that the specimens were collected at a single institution, and the number of cases was small. In the future, it will be necessary to collect specimens from other institutions and construct a more robust classification model. Thus, the practical effectiveness should be evaluated via external validation. In this paper, we show results of various experiments on AD MIL, and processed them by supervised learning method using the same patch images as a comparison. Considering various learning models have been proposed25,26, it is necessary to compare the results with those, in the future.
In cytological diagnosis, classification of cancer type is required after the identification of the benign and malignant status. We have proposed a method for automatic classification of the histological types of lung cancer based on supervised learning27. In the future, we plan to develop a tissue type classification method based on a weakly supervised learning method.
Conclusion
In this study, we developed a method for classification of cytological images as benign or malignant using AD MIL, and compared its ability with classification by supervised learning. We show that weakly supervised learning with AD MIL was able to reach the acceptable accuracy obtained with supervised learning and visualize the regions that contributed to the decision by the attention mechanism. We conclude that our method automatically classifies cytological images more easily and accurately than methods based on supervised learning.
References
American Cancer Society. Cancer facts and figures 2015 (accessed 20 December 2020); https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2015/cancer-facts-and-figures-2015.pdf.
Johnston, W. W. Cytologic diagnosis of lung cancer. Principles and problems. Pathol. Res. Pract. 181, 1–36 (1986).
Fujita, H. AI-based computer-aided diagnosis (AI-CAD): The latest review to read first. Radiol. Phys. Technol. 13, 6–19 (2020).
Matsubara, N., Teramoto, A., Saito, K. & Fujita, H. Bone suppression for chest X-ray image using a convolutional neural filter. Phys. Eng. Sci. Med. 43, 97–108 (2019).
Wang, X., Peng, Y., Lu, Z. L., Bagheri, M. & Summers, R. ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In IEEE CVPR 3462–3471 (2017).
Teramoto, A., Fujita, H., Yamamuro, O. & Tamaki, T. Automated detection of pulmonary nodules in PET/CT images: Ensemble false-positive reduction using a convolutional neural network technique. Med. Phys. 43, 2821–2827 (2016).
Yan, K., Wang, X., Lu, L. & Summers, R. M. DeepLesion: Automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J. Med. Imaging 5, 036501 (2018).
Onishi, Y. et al. Investigation of pulmonary nodule classification using multi-scale residual network enhanced with 3DGAN-synthesized volumes. Radiol. Phys. Technol. 13, 160–169 (2020).
Zhou, X., Takayama, R., Wang, S., Hara, T. & Fujita, H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med. Phys. 44, 5221–5233 (2017).
Bándi, P. et al. From detection of individual metastases to classification of lymph node status at the patient level: The CAMELYON17 challenge. IEEE Trans. Med. Imaging 38, 550–560. https://doi.org/10.1109/TMI.2018.2867350 (2019).
Gao, Y., Davis, J. E. & Saltz, J. H. Patch-based convolutional neural network for whole slide tissue image classification. In IEEE CVPR 2424–2433 (2016).
Iizuka, O. et al. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci. Rep. 10, 1504. https://doi.org/10.1038/s41598-020-58467-9 (2020).
Sirinukunwattana, K. et al. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 35, 1196–1206 (2016).
Teramoto, A. et al. Automated classification of benign and malignant cells from lung cytological images using deep convolutional neural network. Inform. Med. Unlocked 16(1–7), 100205 (2019).
Carbonneau, M.-A., Cheplygina, V., Granger, E. & Gagnon, G. Multiple instance learning: A survey of problem characteristics and applications. Pattern Recognit. 77, 329–353 (2018).
Chaudhari, S., Mithal, V., Polatkan, G. & Ramanath, R. An attentive survey of attention models. J. ACM. 37, 4 article 111 (2020).
Hashimoto, N. et al. Multi-scale domain-adversarial multiple-instance CNN for cancer subtype classification with unannotated histopathological images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 3852–3861 (2020).
Ilse, M., Tomczak, J. & Welling, M. Attention-based deep multiple instance learning. In Proceedings of the 35th International Conference on Machine Learning, PMLR 80 2127–2136 (2018).
LeCun, Y., Bottou, L., Bengio, Y. & Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324 (1998).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. Commun. ACM Proc. Neural Inf. Process. Syst. 60, 84–90 (2017).
Szegedy, C. et al. Going deeper with convolutions. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 07–12-June-2015, 1–9 (2015).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, Nevada 2016 770–778 (2016).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In Proceedings of the 30th IEEE Conference on Computer Vision and PATTERN Recognition, CVPR 2017 2017-January, 2261–2269 (2017).
Kingma, D. & Adam, B. J. A method for stochastic optimization; 2017:ArXiv.
Wang, X. et al. UD-MIL: Uncertainty-driven deep multiple instance learning for OCT image classification. IEEE J. Biomed. Health Inform. 24, 3431–3442 (2020).
Wang, S. et al. RMDL: Recalibrated multi-instance deep learning for whole slide gastric image classification. Med. Image Anal. 58, 101549 (2019).
Teramoto, A., Tsukamoto, T., Kiriyama, Y. & Fujita, H. Automated classification of lung cancer types from cytological images using deep convolutional neural networks. BioMed Res. Int. 2017, 1–6 (2017).
Author information
Authors and Affiliations
Contributions
Conceptualization, A.T, T.T., K.S., and H.F.; Data curation, Y.K., E.S., A.M., and K.I.; Investigation, A.T., T.T., and H.F.; Methodology, A.T.; Project administration, A.T. and K.S.; Software, A.T.; Validation, A.T., T.T., Y.K., E.S., and A.M.; Visualization: A.T.; Writing—Original Draft Preparation, A.T.; Writing—review and editing, T.T., K.S., and H.F. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Teramoto, A., Kiriyama, Y., Tsukamoto, T. et al. Weakly supervised learning for classification of lung cytological images using attention-based multiple instance learning. Sci Rep 11, 20317 (2021). https://doi.org/10.1038/s41598-021-99246-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-021-99246-4
This article is cited by
-
Not seeing the trees for the forest. The impact of neighbours on graph-based configurations in histopathology
BMC Bioinformatics (2025)
-
Automatic lung cancer subtyping using rapid on-site evaluation slides and serum biological markers
Respiratory Research (2024)