Abstract
Wolbachia is one of the most abundant intracellular symbionts of arthropods and has profound effects on host biology. Wolbachia transmission and host phenotypes often depend on its density within the host, which can be affected by multiple biotic and abiotic factors. However, very few studies measured Wolbachia density in natural host populations. Here, we describe Wolbachia in the pear psyllid Cacopsylla pyri from three populations in the Czech Republic. Using phylogenetic analyses based on wsp and multilocus sequence typing genes, we demonstrate that C. pyri harbours three new Wolbachia strains from supergroup B. A fourth Wolbachia strain from supergroup A was also detected in parasitised immatures of C. pyri, but likely came from a hymenopteran parasitoid. To obtain insights into natural Wolbachia infection dynamics, we quantified Wolbachia in psyllid individuals from the locality with the highest prevalence across an entire year, spanning several seasonal generations of the host. All tested females were infected and Wolbachia density remained stable across the entire period, suggesting a highly efficient vertical transmission and little influence from the environment and different host generations. In contrast, we observed a tendency towards reduced Wolbachia density in males which may suggest sex-related differences in Wolbachia-psyllid interactions.
Similar content being viewed by others
Introduction
Wolbachia is one of the most abundant intracellular symbionts of nematodes, insects and other arthropods1,2. It is particularly well-represented among terrestrial arthropod species, with 40–60% of all species estimated to be infected with Wolbachia1,3. Wolbachia has strong impacts on host biology, ecology and evolution, including manipulation of host reproduction via male-killing, feminization, parthenogenesis or cytoplasmatic incompatibility (CI) to promote its transmission4,5,6,7,8. On the other hand, there is strong evidence that Wolbachia acts as a nutritional mutualist in certain species, providing its host with vitamins and nutrients that are important for host survival9,10,11,12,13. Sixteen genetically distinct evolutionary lineages of Wolbachia (supergroups) have been found so far, which differ from each other in host range and biology14,15,16,17. The majority of the known Wolbachia strains fall into two major supergroups A and B, mostly consisting of reproductive parasites.
Wolbachia is mainly vertically transmitted from mother to offspring but can also be transmitted horizontally to new host lineages and infect populations of both related and unrelated host taxa16,17,18,19,20,21,22. Moreover, Wolbachia is used to limit the transmission of numerous human pathogens and agricultural pests13,23,24,25,26,27,28, which makes Wolbachia a promising tool for controlling vector-borne diseases29,30,31. Thus, several Wolbachia strains showed their efficiency in inhibiting mosquito-borne diseases when introduced into natural mosquito populations23,32,33,34. Likewise, several studies on agricultural pests suggested that Wolbachia affects the transmission of plant pathogens24,26,27,35,36,37,38.
While Wolbachia’s influence on their hosts was intensively investigated in the last two decades, e.g.4,5,8,39,40, factors that might affect its density dynamics were less often considered. Maintaining an optimal density is crucial for Wolbachia to ensure long-term and stable relationships with its host8,41. Hence, a reduced infection density may impede the vertical transmission of Wolbachia, while an excessive bacterial density could be detrimental for the host42. Moreover, many Wolbachia-induced phenotypes (reproductive manipulation, protection against pathogens) depend on sufficient Wolbachia density43. For instance, the most frequently observed Wolbachia-induced reproductive phenotype (CI) results in embryonic death when Wolbachia-infected males reproduce with uninfected females4,8. Hence, sufficient Wolbachia density in males is required to induce CI, while females are under selective pressure to maintain a sufficient Wolbachia concentration to protect themselves against CI. Nonetheless, it has also been observed that Wolbachia can be suppressed in males, potentially to reduce CI-related mortality when mating with uninfected females4,5,6,7,8. Environmental conditions (e.g. temperature, humidity), host factors (e.g. host longevity, ageing, genetic background) and interactions with other microorganisms can also have considerable effects on Wolbachia titer41,44,45,46,47,48. The importance of host genotype and Wolbachia strain for the regulation of Wolbachia density was demonstrated in fruit flies, parasitoid wasps, beetles, mosquitoes and isopods49,50,51,52,53,54. Competition for resources and space as well as interactions with other microorganisms, including viruses, bacteria and different Wolbachia strains, can also lead to changes in Wolbachia titer55,56,57. Wolbachia density can also fluctuate during the lifespan of a host, with varying outcomes depending on the host species. While the density declined with age in flies and Aedes mosquitoes58,59,60, it increased in aged Culex pipiens mosquitoes61. Similarly, a recent study on the flies Drosophila simulans and D. melanogaster demonstrated that their respective Wolbachia strains varied differently with host age: while Wolbachia density decreased in ageing males of D. simulans, its density in D. melanogaster increased62. These differences might be linked to differences in host immune response to Wolbachia infection62. In addition, the host mating pattern affected Wolbachia density dynamics in mites63.
The effect of temperature is especially important for Wolbachia density41,45,51,64,65. Depending on the Wolbachia-host association, high temperatures eliminated Wolbachia from their hosts64,66,67,68, increased the Wolbachia titer69or reduced the efficiency of Wolbachia transmission in in vitro experiments70. Thermal sensitivity of Wolbachia abundance in the fly Drosophila melanogaster on a global scale was recently demonstrated71. In contrast to experiments under controlled laboratory conditions, environmental factors such as temperature are much more variable in the wild, making it difficult to translate patterns from laboratory experiments to natural populations. Very few studies investigated the seasonal variations of Wolbachia infection in natural host populations25,72,73,74,75. Of these, only one study73 measured Wolbachia abundance using quantitative PCR (qPCR), and demonstrated a decrease of Wolbachia density across several generations of the pale grass blue butterfly, Pseudozizeeria maha, from early summer to autumn in Japan. The other studies on Wolbachia dynamics in natural host populations25,72,73,74,75 made their conclusions based on the relative abundance of Wolbachia in metabarcoding data. As a result, we are currently lacking insights into the dynamics of Wolbachia infections in natural host populations, particularly for non-model organisms.
Wolbachia was detected in many psyllid species (Insecta: Hemiptera: Psylloidea)76,77,78,79,80 but it was thoroughly studied only in pest species, such as the citrus psyllid Diaphorina citri and the potato psyllid Bactericera cockerelli. Particularly, the studies on D. citri suggest that Wolbachia could mitigate and even impede the transmission of the pathogen ‘Candidatus Liberibacter asiaticus’35,37,81,82. In addition, several Wolbachia strains from B. cockerelli are able to induce CI83,84,85,86. The recent metabarcoding study by87 revealed high relative abundance of Wolbachia in several populations of the pear psyllid Cacopsylla pyri (Psyllidae) from the Czech Republic. In one of these populations, Wolbachia was detected across different developmental stages and seasonal generations throughout an entire year.
In the present study, we aim to investigate Wolbachia infection and density dynamics throughout an entire year in natural populations of C. pyri. This psyllid is an economically important pest that transmits ‘Candidatus Phytoplasma pyri’, the causative agent of the Pear Decline disease of pear trees in Europe88. In contrast to many other psyllid species from northern temperate latitudes, C. pyri is a non-migrating species that usually spends its entire life cycle on pear trees and therefore it is present in pear orchards all year long, producing several summer generations and one overwintering generation89,90. In Central Europe, C. pyri starts reproducing in the middle or at the end of spring, giving rise to the first summer generation after overwintering on its host-plant89. This makes C. pyri a suitable model system to characterise Wolbachia seasonal dynamics in a natural environment. In the current study, we (i) characterise Wolbachia in C. pyri from three localities from the Czech Republic, (ii) analyse the phylogenetic relationships between the different Wolbachia strains and (iii) investigate the density dynamics of the most prevalent Wolbachia strain in one population throughout an entire year using qPCR. To our knowledge, this is the first study demonstrating Wolbachia density dynamics in a natural host population across an entire year using a targeted qPCR approach. Our results provide new insights into the impact of seasonal dynamics on Wolbachia density in a natural population of psyllids and will broaden our general understanding of Wolbachia-host interactions in natural environments.
Results
Wolbachia prevalence
From the 56 analysed individuals of C. pyri (Fig. 1a), 38 were tested positive for Wolbachia (Table 1). The highest Wolbachia prevalence was observed in the population CZ2 (94.4%; N = 34/36), while in CZ1 (N = 1/10) and CZ3 (N = 1/5) the Wolbachia prevalence reached only 10% and 20%, respectively (Fig. 1b). Moreover, the two out of five (40%) parasitised immatures from CZ1 were infected with Wolbachia.
(a) Adult pear psyllid Cacopsylla pyri (photograph by O. Michálek). (b) Wolbachia prevalence in each of three studied populations of C. pyri in the Czech Republic. Pie charts represent the percentage of C. pyri individuals infected with different Wolbachia strains (wCpyr1, wCpyr2, wCpyr3 and wSaph) or not infected with Wolbachia. Sample size per population is indicated by the size of the pie chart. Details of localities and sample size are given in Table 1. The map was produced in ArcGIS Desktop v10.8.2 (ESRI) (https://desktop.arcgis.com).
Identification of Wolbachia strains and supergroups in Cacopsylla pyri
Sanger sequencing of wsp and MLST genes was performed for all 38 Wolbachia-infected individuals, except for the ftsZ gene of individuals from CZ3 which failed to amplify. Sequence analysis indicated the presence of three novel alleles for fbpA, two novel alleles for wsp and ftsZ, and one for coxA, gatB and hcpA (Table 2). Together, these alleles represented four new Wolbachia sequence types (Table 2, Supplementary Table S1). Chromatograms showed clear single peaks, suggesting the presence of just one Wolbachia strain in each individual, which allowed to assign the different alleles to three different strains. They were named wCpyr1, wCpyr2 and wCpyr3. None of the strains were previously described in other organisms.
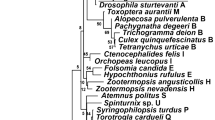
Both ML and BI phylogenetic analyses based on the wsp and MLST genes showed that all three Wolbachia strains found in C. pyri belong to supergroup B (Fig. 2, Supplementary Fig. S1). The strains wCpyr1 and wCpyr2 were differentiated by a single nucleotide in the fbpA gene (p-distance = 0.003), whereas the strain wCpyr3 was more divergent from both wCpyr1 and wCpyr2 for three of the five analysed genes (wsp: p-distance = 0.144; coxA: p-distance = 0.148; fbpA: p-distance = 0.065 and 0.064, respectively; gatB: p-distance = 0.003; hcpA: p-distance = 0.002) (Supplementary Table S1). MLST alleles gatB and hcpA of wCpyr1 and wCpyr2 showed 100% identity with the strain wNriv1 from the butterfly Neptis rivularis (Table 2, Supplementary Table S1). Similarly, BLAST identified identical sequences to gatB and hcpA of wCpyr1 and wCpyr2 in several other strains. Among them, gatB sequences were identical to wDec from the mosquito Culex decens and wOscaB from the butterfly Ostrinia scapulalis; hcpA sequences were identical to wCon from the beetle Tribolium confusum and an unnamed strain from the froghopper Philaenus spumarius. In contrast, wsp, fbpA and ftsZ of wCpyr1 and wCpyr2 represented new alleles based on comparisons with both the MLST database and BLAST. In spite of the aforementioned similarities in two MLST genes, the wsp sequences of wDec, wOscaB and wCon were quite different from the wsp of wCpyr1 and wCpyr2 with a genetic divergence of 0.167, 0.209 and 0.194, respectively. Based on the comparisons of p-distances, gatB demonstrated the least sequence divergence, ranging from 0.001 to 0.010. These differences between the wsp and the MLST genes were also reflected in the phylogenetic trees: according to the wsp tree, wCpyr1 and wCpyr2 were most closely related to two strains from Drosophila simulans (wNo, wMa) and two strains from the mosquitoes Aedes albopictus (wAlbB) and Culex quinquefasciatus (wPip) (Fig. 2a, Supplementary Fig. S1a).
Maximum likelihood tree of Wolbachia (a) wsp gene sequences, (b) concatenated MLST gene sequences. Bootstrap values > 60% are shown. The new strains wSaph, wCpyr1, wCpyr2 and wCpyr3 are highlighted. Wolbachia supergroups are indicated by the coloured bar on the right-hand side. GenBank accession numbers for the strains used for the phylogenetic analyses are provided in the Supplementary Table S3.
Based on comparisons with alleles from the MLST database, four genes (wsp, coxA, fbpA and gatB) of wCpyr3 were identical to those of the strain wEjen from the butterfly Erebia jeniseiensis (Table 2, Supplementary Table S1). Additionally, BLAST searches showed that gatB of wCpyr3 was identical to several other Wolbachia strains, including wEbl from the butterfly Eurema blanda and wB from the parasitoid wasp Megastigmus pistaciae, and fbpA of wCpyr3 was identical to wAlbB from the mosquito Aedes albopictus. In contrast to wsp, coxA, fbpA and gatB, we found no sequences identical to the hcpA allele of wCpyr3. Similar to the results on the allele comparison, the ML and BI wsp trees strongly supported the clade with wCpyr3 and the two closely-related strains wB and wEjen within supergroup B (BS = 91; BI = 1) (Fig. 2a, Supplementary Fig. S1a).
Additional ML and BI phylogenetic analyses were performed including also the wsp sequences of previously identified Wolbachia strains from other psyllid species. The results demonstrated that the Wolbachia strains from C. pyri belonged to different clades from the strains of other psyllid species, including those from other Cacopsylla species (Supplementary Fig. S2). Nevertheless, the Wolbachia strains from all analysed psyllid species belong to supergroup B.
Identification of a Wolbachia strain of potentially parasitoid origin
Two of the five immature individuals of C. pyri from CZ1 were infected with another Wolbachia strain that was placed within supergroup A (wsp: BS = 93, BI = 1; MLST: BS = 66, BI = 1) (Figs. 2a–b, Supplementary Figs. S2a–b, Table 1). These immatures had been shown to be parasitised according to a COI barcoding approach87. Although the identification of the parasitoid present in the immatures was not formal, the detected Wolbachia strain was named wSaph after its potential host, S. aphidivorus. Based on comparisons with the MLST database, the wsp and MLST alleles (except hcpA) of wSaph were different from previously sequenced strains present in this database (Table 2, Supplementary Table S1). Additionally, BLAST searches revealed that the hcpA gene of wSaph was identical to several other strains, including wBif from the fly Drosophila bifasciata and wRha from the parasitoid wasp Psyttalia carinata (Braconidae), whereas fbpA of wSaph was identical to wBif.
Density dynamics of wCpyr2 throughout an entire year
The C. pyri population CZ2 had the highest Wolbachia prevalence among the studied populations (94.4%; Fig. 1b) and therefore was chosen for monitoring of the Wolbachia density dynamics. To investigate whether seasonal changes of local abiotic (Supplementary Fig. S3) and biotic factors affected the density of Wolbachia in C. pyri, we compared the densities of wCpyr2 from psyllid individuals sampled from CZ2 throughout the year. Among all quantified psyllid specimens (N = 36), only two males (P58 and P61; Supplementary Table S2) collected in November and December, were negative. Hence, either these individuals were not infected with wCpyr2 or this strain was present at low titers not detectable with our qPCR approach (Fig. 3a). Psyllid individuals sampled in November and December were excluded from the statistical analysis as only two infected individuals remained for these months after eliminating the two uninfected males from the dataset.
Seasonal dynamics of Wolbachia (wCpyr2) density in Cacopsylla pyri individuals (a) throughout the sampling year, (b) between immatures, females and males. The shaded areas correspond to the period of occurrence of the overwintering generation of C. pyri, the clear area represent the period of occurrence of its summer generations. For more details on the analysed individuals of C. pyri, see Supplementary Table S2.
Monitoring of wCpyr2 titers throughout the remaining eight months revealed that Wolbachia titer/host cell in all individuals ranged between 0.06 and 16.91 (Supplementary Table S2). The individuals collected in May showed a significantly lower titer (mean Wolbachia titer/host cell: 0.42) compared to individuals sampled in all other months (mean Wolbachia titer/host cell: 1.62 in February—16.91 in June) (Tukey’s post-hoc test: p ≤ 0.0002). When considering also the sex of the tested individuals, our data showed that the Wolbachia titer in females was relatively stable throughout the year, while it was more variable in males (Figs. 3a–b). In May, only males were captured and those had the lowest Wolbachia titers detected during the entire study period, thereby driving this drop in Wolbachia titer in May. Despite a tendency towards lower Wolbachia titers in males, a global comparison of the Wolbachia titers between males and females across all time points was not significant (Welsh’s t-test: t = 2.2067, df = 9.6116, p = 0.0529) (Fig. 3b). This outcome may have been influenced by the low number of males (N = 12) collected only in five out of ten sampling months (Supplementary Table S2). In addition, two immatures collected in May also had low Wolbachia titers (0.06 and 0.30 Wolbachia/host cell) (Fig. 3b), but these individuals were not included in the statistical analysis due to their different developmental stage.
Discussion
Wolbachia density plays an important role in the evolution of symbiotic associations and ecological interactions, mainly through its impact on symbiont transmission and host extended phenotype8,41,50,52,66,71. Although different environmental factors have a strong influence on Wolbachia titre64,66,67,68,70,91, the majority of studies focusing on Wolbachia density dynamics were performed under controlled laboratory conditions. Given that host-microbe associations in the wild are likely influenced by multiple environmental factors (e.g. temperature, humidity), host factors (e.g. host longevity, ageing, genetic background) and interactions with other organisms, controlled laboratory environments cannot encompass the full spectrum of these factors and their potential impacts41,44,45,46,47,48,92. In the current study, we present one of the few examples of the dynamics of Wolbachia in a natural host population across different generations and seasons.
We assessed the Wolbachia density dynamics in a natural population of the pear psyllid C. pyri, across several generations throughout an entire year. Cacopsylla pyri completes its life cycle on pear trees and therefore represents an interesting model to study Wolbachia density dynamics through different seasons. This psyllid species produces several summer generations and one overwintering generation on pear trees89,90, which allows studying the impact of varying environmental conditions on the Wolbachia density throughout the year. Only a few studies attempted to describe Wolbachia seasonal dynamics in natural host populations so far72,73,74. In the butterfly Pseudozizeeria maha, the Wolbachia density decreased across several host generations from spring/early summer to autumn73. Likewise, the relative abundance of Wolbachia within the microbiome declined from spring to summer in the flea Synosternus cleopatrae74. A decrease in Wolbachia relative abundance, through its vertical transmission to the subsequent host generation, was correlated with higher temperatures in natural populations of the mosquito complexes Aedes vexans and Culex pipiens/restuans25. However, as none of these studies included individuals sampled in winter, the effect of harsh winter conditions on Wolbachia dynamics in overwintering hosts has not been analysed yet.
To investigate Wolbachia density dynamics in a natural population of C. pyri across all seasons, we measured the titer of the most frequently occurring strain (wCpyr2) at a single locality throughout an entire year. Although the bacterial density in natural populations can be influenced by multiple factors, Wolbachia density was stable in the analysed females of C. pyri, which were abundantly infected with Wolbachia throughout the sampling period, suggesting a highly efficient vertical transmission of wCpyr2 from mothers to daughters across different host generations. Regardless of their fluctuations (e.g. a significant decrease of local temperature during the winter, Fig. S3), the environmental conditions had no impact on Wolbachia density in females. This might indicate a strong selective pressure on females to maintain a sufficient Wolbachia density in order to protect themselves against cytoplasmatic incompatibility (CI)4. As such, several Wolbachia strains from supergroup B that are closely related to wCpyr2 are known to induce CI, i.e. two strains from the fly Drosophila simulans (wMa, wNo) and two strains from the mosquitoes Aedes albopictus (wAlbB) and Culex quinquefasciatus (wPip)93,94,95. Taken together, this suggests that wCpyr2 might also be able to induce CI but this needs to be validated experimentally.
In contrast to females, not all tested males harboured Wolbachia and its titer in males was more variable compared to the females. Considering the efficient Wolbachia transmission from mothers to daughters, it is unlikely that male embryos received less Wolbachia from their mothers. Instead, Wolbachia could multiply less in males or could be suppressed in males in order to reduce CI-related mortality when mating with an uninfected female. This would result in reduced Wolbachia titers in aged males. Such a pattern was predicted by population genetic models and observed in the mosquito Aedes albopictus and the fly Drosophila innubila58,59,96. In accordance with these predictions, male individuals in our study showed a general tendency towards reduced Wolbachia density compared to females, culminating in a significant drop in Wolbachia titer in May. However, since no females were sampled in May, we cannot exclude that the titer might have been low also in females at this time point. In Central Europe, a new annual cycle of C. pyri begins in April–May, giving rise to the first summer generation after overwintering and newly-emerged adults from this generation can be already found in the second half of May89. At this time, aged adults from the previous overwintering generation can still be alive and present in the orchard as well. In contrast to D. innubila and A. albopictus mosquitoes, the Wolbachia titer of mosquitoes from the Culex pipiens group increased with ageing and therefore young males of C. pipiens had a lower Wolbachia titer compared to older individuals61. Consequently, the males of C. pyri collected in May with a low Wolbachia titer could have been either old individuals from the overwintering generation (if Wolbachia titer decreases with age) or young summer-form adults (if Wolbachia titer increases with age). Both scenarios would agree with those from the previous studies that demonstrated the impact of age on Wolbachia titer in laboratory studies of different systems58,59,60,61,62. Moreover, the number of males collected throughout the study period was half the number of females. While this could be a coincidence, it could also be the result of another Wolbachia-induced reproductive manipulation, e.g. male-killing or feminization4,5,6,7,8. However, the presence of Wolbachia in males, albeit at more variable titer, makes this scenario less likely.
In the current study, we characterised not only wCpyr2 but also two additional Wolbachia strains (wCpyr1 and wCpyr3) which all belong to Wolbachia supergroup B. Each strain was detected as a single infection in individuals from three different localities in the Czech Republic. Hence, multiple Wolbachia strains occur in this host species, but they appear not to co-occur in a same individual of C. pyri. Currently, we cannot draw any conclusions whether the observed patterns of Wolbachia infection reflect stable host-symbiont associations with at least two divergent strains (e.g.97), an ongoing spread of one or more newly-acquired Wolbachia strains (e.g.98), or replacement of a pre-existing strain by a new, more competitive strain (e.g.99). To predict Wolbachia infection spread and potential introductions into new host populations, further studies should investigate the infection dynamics of the newly discovered Wolbachia strains on a larger geographical scale. In future studies, it is also necessary to sequence the whole genome of the new Wolbachia strains to understand how they diverged. This is particularly relevant for the strains wCpyr1 and wCpyr2 that differed from each other only slightly based on the MLST genes. Wolfe et al.100 demonstrated that small differences in the MLST genes can in fact reflect bigger structural differences in the whole genome of the symbiont. Additionally, it was striking to find that neither of the strains wCpyr1, wCpyr2 and wCpyr3 showed close relatedness to the Wolbachia strains from other psyllid species, including other Cacopsylla species (e.g.79).
An additional Wolbachia strain was detected in immatures of C. pyri parasitised by a hymenopteran wasp that was tentatively identified as Syrphophagus aphidivorus based on insect DNA barcoding. Syrphophagus spp. are relatively frequent parasitoids or hyperparasitoids of various psyllids, including Cacopsylla spp. on pear trees101. This Wolbachia strain clustered within supergroup A and was closely related to Wolbachia strains from other hymenopteran parasitoid wasps. Despite a high importance of parasitoids for a biological control of psyllids, only limited information is available on the microbial communities of their parasitoids102,103. Particularly, Wolbachia was recently detected in the parasitoids of the citrus psyllid Diaphorina citri103, whereas to our knowledge, our study represents the first detection of Wolbachia in parasitised individuals of Cacopsylla. Considering the high incidence of parasitoids in pear psyllid populations, particularly in immatures104,105, horizontal transmission of Wolbachia between psyllids via shared parasitoids might occur, as it was described in other host-parasitoid associations18,106,107,108. However, a horizontal transmission from parasitoids to their hosts is quite unlikely, as parasitised hosts die and therefore are not able to maintain the bacteria throughout later life stages and pass them onto next generations1,19,102. Thus, it is more likely that bacterial symbionts hitchhike from hosts to parasitoids. In the case of wSaph, we can only speculate whether this Wolbachia strain originally belonged to the parasitoid or to C. pyri. To investigate the possibilities of horizontal transmission of wSaph in the wild, further experimental studies on host-parasitoid interactions in pear orchards are required.
In the current work, we studied the density dynamics of the newly-described Wolbachia strain wCpyr2 of the pear psyllid C. pyri across an entire year, spanning several host generations. We demonstrated that Wolbachia density remained stable in females, while we observed a tendency towards reduced Wolbachia density in males, with a significant drop in Wolbachia titer in May. However, in light of a relatively low sample size of C. pyri, these results should be interpreted with caution. Moreover, in similar studies we highly recommend to do the non-destructive DNA extractions and retain voucher specimens in order to be able to identify the generations of the analysed individuals. Nevertheless, our study is the first demonstration of the seasonal dynamics of Wolbachia in psyllids and the first study to monitor Wolbachia abundance in a natural host population across an entire year. The identification of three different Wolbachia strains in C. pyri and another strain in parasitised individuals of C. pyri highlight that additional studies are needed to study phenotypic effects and spatial dynamics of Wolbachia in natural host populations of this important pest species. Taken together, this study provides insights into the ecological and evolutionary dynamics of Wolbachia-host interactions in the natural environment and opens new research questions on this study system.
Methods
Psyllid sampling
Adult and immature specimens of Cacopsylla pyri (Fig. 1a) were collected from three different localities in the Czech Republic: Litenčice = CZ1, Starý Lískovec (Brno) = CZ2, and Staré Město (Svitavy) = CZ3 (Fig. 1b, Table 1). In the locality CZ2, C. pyri adults were collected throughout an entire year, from February 2020 to February 2021, with a time interval of 7–10 days between each visit. In contrast, only one sampling was performed in CZ1 and CZ3. At each visit, psyllids were collected from five branches of five randomly chosen pear trees. Adult specimens of C. pyri were sampled using entomological sweep nets and a beating tray, while 4–5th instar immatures were collected with a camelhair brush. All individuals were immediately stored in absolute ethanol and later kept at − 20 °C. Cacopsylla pyri was identified based on the morphological keys by109,110.
Several immatures of C. pyri from CZ1 were shown to be parasitised based on COI gene sequences amplified from the sample DNA87. This barcoding approach allowed a tentative identification of the parasitoid as Syrphophagus aphidivorus (Hymenoptera: Encyrtidae) based on the closest BLAST hit. The immatures of C. pyri from CZ1 were included in the current study to explore Wolbachia infection in parasitised psyllid individuals.
DNA extraction and strain identification
Fifty-six individuals (17 ♂, 27 ♀, 12 imm.) of C. pyri from the three aforementioned populations, CZ1 (N = 15), CZ2 (N = 36) and CZ3 (N = 5), were tested for Wolbachia infection (Table 1). The DNA of 43 individuals (14 ♂, 24 ♀, 5 imm.) was extracted using the DNeasy Blood and Tissue Kit (Qiagen), while the DNA of 13 specimens (3 ♂, 3 ♀, 7 imm.), included in our previous metabarcoding study87, had been extracted using E.Z.N.A.® Tissue DNA Kit (Omega Bio-tek).
Identification of Wolbachia strains and supergroups was based on the amplification of the Wolbachia surface protein wsp111 and the five multilocus sequence typing (MLST) genes coxA, fbpA, ftsZ, gatB and hcpA112. Due to its fast-evolving rate, wsp provides many informative characters that help resolving the relationships between different Wolbachia strains113,114. The MLST genes are more conserved and were broadly applied for Wolbachia strain detection in numerous studies (e.g14,15,17,114,115,116,117). Each 25 μl PCR reaction was composed of 1X DreamTaq PCR Master Mix (Thermo Fisher), 7 μl of sterile water, 0.7 μM of each primer and 2 μl of DNA template. PCR conditions were as follows: 95 °C for 5 min; 40 cycles of 95 °C for 15 s, the optimal annealing temperature (see below) for 30 s and 72 °C for 1 min; 72 °C for 10 min. The optimal PCR annealing temperature was 53 °C for hcpA, 54 °C for both gatB and ftsZ, 55 °C for coxA, and 59 °C for fbpA112. PCR conditions for wsp were as follows: 95 °C for 5 min; 40 cycles of 95 °C for 15 s, 58 °C for 30 s and 72 °C for 1 min; 72 °C for 10 min, using the primer pair 81F and 691R111. PCR products were purified and Sanger sequenced by Eurofins Genomics (Ebersberg, Germany). Each PCR product was sequenced in both directions and the consensus sequences were obtained using ‘MEGA X’118.
Phylogenetic characterization of Wolbachia strains based on wsp and MLST
The wsp sequences were aligned with the 21 most closely related Wolbachia strains (Supplementary Table S3) identified from both the Wolbachia MLST database119 and GenBank based on BLAST120. The alignment (Supplementary File 1) was produced using the MAFFT v7 web server, applying the G-INS-I strategy121. Additionally, an extended wsp tree was produced including the wsp sequences from other psyllid-associated Wolbachia strains from GenBank as well (Supplementary File 2). The five MLST genes were concatenated (1970 bp, Supplementary File 3) and manually aligned with the same 21 Wolbachia strains (Supplementary Table S3) using ‘MEGA X’118. The phylogenetic relationships were evaluated through maximum likelihood (ML) analyses using IQ-TREE v1.6.12122 and Bayesian Inference (BI) analysis using MrBayes v3.2.7a123. As an outgroup, we used the Wolbachia strain of Folsomia candida from supergroup E (Supplementary Table S3). Nodal support for the ML analysis was calculated using a standard nonparametric bootstrap (BS) with 1000 replicates. Clades with BS > 70% were considered strongly supported and with BS 60–70% moderately supported. For the BI analysis, the best substitution model for each gene partition was estimated using Jmodeltest v2.1.10124 based on Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) scores (Supplementary File 4). The analysis of 15 million generations was run on the CIPRES platform125. Nodal support was assessed by posterior probabilities (PP). Nodes were considered strongly supported with PP > 90%. Both ML and BI trees were visualized with iTOL v6126.
Consensus sequences obtained in this study were submitted to GenBank with the following accession numbers: ON146571–ON146574 and ON157498–ON157516 (Supplementary Table S3).
Quantitative PCR and statistical analyses
In total, for the quantitative PCR we analysed 36 specimens (12 ♂, 22 ♀, 2 imm.) of C. pyri collected from CZ2 across an entire year from February 2020 to February 2021. At least three psyllid individuals were captured for each month of the sampling year, with the exception of March and September, when no C. pyri individuals were found.
All samples were run in duplicates on a CFX96 real-time PCR system (Bio-Rad, Hercules, CA, USA). Wsp and the host gene wingless (wg) were amplified using primers specific for the analysed Wolbachia strain wCpyr2 and the wg gene of C. pyri. For wsp, the primer set from Le Clec'h et al.127 was modified, producing a 202 bp-amplicon: wCpyr_Fq (5'-TGGTGCAGCATTTACTCCAAC-3') and wCpyr_Rq (5'-TTGCTTGATAAGCAAAACC-3'). PCR conditions were as follows: 95 °C for 3 min; 40 cycles of 95 °C for 15 s, 55 °C for 30 s and 72 °C for 30 s. For wg, we designed new primers amplifying a 186 bp amplicon: Wg-202Fq (5'-CTCGTCTACCTGGAGACCTC-3') and Wg-362Rq (5'-ACGCAGGAAATCACTGTT-3'). PCR was performed with the following conditions: 95 °C for 3 min; 40 cycles of 95 °C for 15 s, 58 °C for 30 s and 72 °C for 30 s. Each 20 μl reaction contained 1X IQ SYBR Green Supermix (Bio-Rad), 6 μl of sterile water, 0.5 μM of each primer and 2 μl of DNA template. The amplification efficiency of the wsp and wg primer pairs was tested using a standard curve at different annealing temperatures to determine the optimal annealing temperature for the highest amplification efficiency, which was 92% for wsp and 95% for wg. To verify the amplification of the target PCR product across all reactions, a melting curve analysis was performed at the end of each run. Gene copy numbers were determined based on standard curves consisting of tenfold serial dilutions of longer PCR products of the same genes. The longer PCR products for the standard curves were amplified by standard PCR using the DreamTaq PCR Master Mix (Thermo Fisher) and the following primer sets: 81F and 691R for wsp111 and 19F 5'-ACATGYTGGATGAGAYTACCA-3' and 388R 5'-TCTTGTGTTCTATAACCACGCCCAC-3' for wg (this study). PCR conditions for wg were as follows: 95 °C for 5 min; 40 cycles of 95 °C for 15 s, 58 °C for 30 s and 72 °C for 1 min; 72 °C for 10 min. The resulting amplicons were purified using AMPure XP beads (Beckman-Coulter) and quantified using the Qubit dsDNA High Sensitivity Assay Kit (Invitrogen).
To calculate the Wolbachia titer/host cell ratio, the mean number of wsp copies was divided by the mean number of wg copies for each specimen. All qPCR data were log-transformed and analysed in R v3.6.3 using the packages agricolae and car128,129,130. Psyllid specimens that were not infected with Wolbachia as well as immatures (for which only two biological replicates were available), were eliminated from the statistical analyses. The dataset was tested for normality and homogeneity of variance using the Shapiro–Wilk and Levene tests, respectively. Welsh’s t-test was used to test for potential differences in Wolbachia titer between males and females. Analysis of variance (ANOVA) followed by Tukey’s post-hoc test for multiple comparisons was used to test for differences in Wolbachia titer between different months of the year. The data was visualized using the ggplot package in R131. The map showing the representation of different Wolbachia strains in each studied population of C. pyri (Fig. 1b) was produced in ArcGIS Desktop v10.8.2 (ESRI).
Data availability
All data generated or analysed during this study are included in the current paper and in its Supplementary Information Files. The datasets generated during the current study in GenBank with the following accession numbers, ON146571–ON146574 and ON157498–ON157516.
References
Weinert, L. A., Araujo-Jnr, E. V., Ahmed, M. Z. & Welch, J. J. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc. R. Soc. B. 282(1807), 20150249 (2015).
Lefoulon, E. et al. Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ 4, e1840 (2016).
Zug, R. & Hammerstein, P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE 7(6), 38544 (2012).
Werren, J. H., Baldo, L. & Clark, M. E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751 (2008).
Engelstädter, J. & Hurst, G. D. D. The ecology and evolution of microbes that manipulate host reproduction. Annu. Rev. Ecol. Evol. Syst. 40, 127–149 (2009).
Hurst, G. D. D. & Frost, C. L. Reproductive parasitism: Maternally inherited symbionts in a piparental world. Cold Spring Harb. Perspect. Biol. 7(4), a017699 (2015).
Bailly-Bechet, M. et al. How long does Wolbachia remain on board?. Molec. Biol. Evol. 34, 1183–1193 (2017).
Kaur, R. et al. Living in the endosymbiotic world of Wolbachia: A centennial review. Cell Host Microbe 29, 879–893 (2021).
Timmermans, M. J. T. N. & Ellers, J. Wolbachia endosymbiont is essential for egg hatching in a parthenogenetic arthropod. Evol. Ecol. 23, 931–942 (2009).
Hosokawa, T. et al. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Nation. Acad. Sci. USA 107, 769–774 (2010).
Nikoh, N. et al. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc. Nation. Acad. Sci. USA 111, 10257–10262 (2014).
Balvín, O., Roth, S., Talbot, B. & Reinhardt, K. Co-speciation in bedbug Wolbachia parallel the pattern in nematode hosts. Sci. Rep. 8(1), 1–9 (2018).
Manoj, R. R. S., Latrofa, M. S., Epis, S. & Otranto, D. Wolbachia: Endosymbiont of onchocercid nematodes and their vectors. Parasites Vectors 14(1), 1–24 (2021).
Gerth, M., Gansauge, M.-T., Weigert, A. & Bleidorn, C. Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nat. Commun. 5(1), 1–7 (2014).
Brown, A. M. V. et al. Genomic evidence for plant-parasitic nematodes as the earliest Wolbachia hosts. Sci. Rep. 6(1), 1–14 (2016).
Kajtoch, Ł et al. Using host species traits to understand the Wolbachia infection distribution across terrestrial beetles. Sci. Rep. 9(1), 1–15 (2019).
Zimmermann, B. L. et al. Supergroup F Wolbachia in terrestrial isopods: Horizontal transmission from termites?. Evol. Ecol. 35, 165–182 (2021).
Heath, B. D., Butcher, R. D. J., Whitfield, W. G. F. & Hubbard, S. F. Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr. Biol. 9, 313–316 (1999).
Vavre, F., Fleury, F., Lepetit, D., Fouillet, P. & Bouletreau, M. Phylogenetic evidence for horizontal transmission of Wolbachia in host- parasitoid associations. Mol. Biol. and Evol. 16, 1711–1723 (1999).
Jiggins, F. M., Bentley, J. K., Majerus, M. E. N. & Hurst, G. D. D. Recent changes in phenotype and patterns of host specialization in Wolbachia bacteria. Mol. Ecol. 11, 1275–1283 (2002).
Schuler, H. et al. Evidence for a recent horizontal transmission and spatial spread of Wolbachia from endemic Rhagoletis cerasi (Diptera: Tephritidae) to invasive Rhagoletis cingulata in Europe. Mol. Ecol. 22, 4101–4111 (2013).
Sanaei, E., Charlat, S. & Engelstädter, J. Wolbachia host shifts: Routes, mechanisms, constraints and evolutionary consequences. Biol. Rev. 96, 433–453 (2021).
Hoffmann, A. A., Ross, P. A. & Rašić, G. Wolbachia strains for disease control: Ecological and evolutionary considerations. Evol. Appl. 8, 751–768 (2015).
Chu, C.-C., Gill, T. A., Hoffmann, M. & Pelz-Stelinski, K. S. Inter-population variability of endosymbiont densities in the Asian citrus psyllid (Diaphorina citri Kuwayama). Microb. Ecol. 71, 999–1007 (2016).
Nováková, E. et al. Mosquito microbiome dynamics, a background for prevalence and seasonality of west nile virus. Front. Microbiol. 8, 526 (2017).
Krstić, O. et al. Wolbachia infection in natural populations of dictyophara Europaea, an alternative vector of grapevine flavescence dorée phytoplasma: Effects and interactions: Effects of wolbachia on phytoplasma vector. Ann. Appl. Biol. 172, 47–64 (2018).
Gong, J.-T. et al. Stable introduction of plant-virus-inhibiting Wolbachia into planthoppers for rice protection. Curr. Biol. 30, 4837–4845 (2020).
Johnston, K. L. et al. Anti-Wolbachia drugs for filariasis. Trends. Parasit. 37(12), 1068–1081 (2021).
Brelsfoard, C. L. & Dobson, S. L. Wolbachia-based strategies to control insect pests and disease vectors. Asia-Pac. J. Mol. Biol. Biotech. 17, 55–63 (2009).
Arora, A. K. & Douglas, A. E. Hype or opportunity? Using microbial symbionts in novel strategies for insect pest control. J. Insect Phys. 103, 10–17 (2017).
Landmann, F. The Wolbachia Endosymbionts. Microbiol. Spectr. 7(2), 72 (2019).
McMeniman, C. J. et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 323, 141–144 (2009).
Bourtzis, K. et al. Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Trop. 132S, S150–S163 (2014).
Schmidt, T. L. et al. Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biol. 15(5), 2001894 (2017).
Fagen, J. et al. Characterization of the relative abundance of the citrus pathogen Ca. Liberibacter Asiaticus in the microbiome of its insect vector, Diaphorina citri, using high throughput 16S rRNA sequencing. Open Microbiol. J. 6, 29–33 (2012).
Kolora, L. D., Powell, C. M., Hunter, W., Bextine, B. & Lauzon, C. R. Internal extracellular bacteria of diaphorina citri Kuwayama (hemiptera: Psyllidae), the Asian citrus psyllid. Curr. Microbiol. 70, 710–715 (2015).
Hosseinzadeh, S. et al. Distribution and variation of bacterial endosymbiont and “Candidatus Liberibacter asiaticus” titer in the huanglongbing insect vector Diaphorina citri Kuwayama. Microb. Ecol. 78, 206–222 (2019).
Moussa, A. et al. Bacterial microbiota associated with insect vectors of grapevine Bois noir disease in relation to phytoplasma infection. FEMS Microbiol. Ecol. 96(11), fiaa203 (2020).
Bordenstein, S. R., O’Hara, F. P. & Werren, J. H. Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature 409, 707–710 (2001).
Johnson, K. The impact of Wolbachia on virus infection in mosquitoes. Viruses 7, 5705–5717 (2015).
López-Madrigal, S. & Duarte, E. H. Titer regulation in arthropod-Wolbachia symbioses. FEMS Microbiol. Let. 366(23), fnz232 (2019).
Min, K.-T. & Benzer, S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Nation. Acad. Sci. 94, 10792–10796 (1997).
Chrostek, E. et al. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: A phenotypic and phylogenomic analysis. PLoS Genet. 9(12), e1003896 (2013).
Stacey, D. A. et al. Genotype and temperature influence pea aphid resistance to a fungal entomopathogen. Physiol. Entomol. 28, 75–81 (2003).
Thomas, M. B. & Blanford, S. Thermal biology in insect-parasite interactions. Trends in Ecol. Evol. 18, 344–350 (2003).
Jaenike, J. Coupled population dynamics of endosymbionts within and between hosts. Oikos 118, 353–362 (2009).
Versace, E., Nolte, V., Pandey, R. V., Tobler, R. & Schlötterer, C. Experimental evolution reveals habitat-specific fitness dynamics among Wolbachia clades in Drosophila melanogaster. Mol. Ecol. 23, 802–814 (2014).
Corbin, C., Heyworth, E. R., Ferrari, J. & Hurst, G. D. D. Heritable symbionts in a world of varying temperature. Heredity 118, 10–20 (2017).
Berticat, C., Rousset, F., Raymond, M., Berthomieu, A. & Weill, M. High Wolbachia density in insecticide–resistant mosquitoes. Proc. R. Soc. Lond. B 269, 1413–1416 (2002).
Mouton, L., Henri, H., Bouletreau, M. & Vavre, F. Strain-specific regulation of intracellular Wolbachia density in multiply infected insects. Mol. Ecol. 12, 3459–3465 (2003).
Mouton, L., Henri, H., Charif, D., Boulétreau, M. & Vavre, F. Interaction between host genotype and environmental conditions affects bacterial density in Wolbachia symbiosis. Biol. Lett. 3, 210–213 (2007).
Kondo, N., Shimada, M. & Fukatsu, T. Infection density of Wolbachia endosymbiont affected by co-infection and host genotype. Biol. Lett. 1, 488–491 (2005).
Duron, O. et al. High Wolbachia density correlates with cost of infection for insecticide resistant Culex pipiens mosquitoe. Evol. 60, 303–314 (2006).
Dittmer, J. et al. Host tissues as microhabitats for Wolbachia and quantitative insights into the bacterial community in terrestrial isopods. Mol. Ecol. 23, 2619–2635 (2014).
Mouton, L. et al. Virulence, multiple Infections and regulation of symbiotic population in the Wolbachia-Asobara tabida symbiosis. Genetics 168, 181–189 (2004).
Goto, S., Anbutsu, H. & Fukatsu, T. Asymmetrical interactions between Wolbachia and Spiroplasma endosymbionts coexisting in the same insect host. Appl. Environ. Microbiol. 72, 4805–4810 (2006).
Mousson, L. et al. Wolbachia modulates Chikungunya replication in Aedes albopictus. Mol. Ecol. 19, 1953–1964 (2010).
Unckless, R. L., Boelio, L. M., Herren, J. K. & Jaenike, J. Wolbachia as populations within individual insects: Causes and consequences of density variation in natural populations. Proc. R. Soc. B. 276, 2805–2811 (2009).
Tortosa, P. et al. Wolbachia age-sex-specific density in Aedes albopictus: A host evolutionary response to cytoplasmic incompatibility?. PLoS ONE 5, e9700 (2010).
Moretti, R. et al. Increased biting rate and decreased Wolbachia density in irradiated Aedes mosquitoes. Paras. Vectors 15(1), 1–16 (2022).
Duron, O., Fort, P. & Weill, M. Influence of aging on cytoplasmic incompatibility, sperm modification and Wolbachia density in Culex pipiens mosquitoes. Heredity 98, 368–374 (2007).
Shropshire, J. D., Hamant, E. & Cooper, B. S. Male age and Wolbachia dynamics: investigating how fast and why bacterial densities and cytoplasmic incompatibility strengths vary. MBio. 12(6), e02998 (2021).
Zhao, D.-X., Zhang, X.-F., Chen, D.-S., Zhang, Y.-K. & Hong, X.-Y. Wolbachia-host interactions: host mating patterns affect Wolbachia density dynamics. PLoS ONE 8(6), e66373 (2013).
Mouton, L., Henri, H., Bouletreau, M. & Vavre, F. Effect of temperature on Wolbachia density and impact on cytoplasmic incompatibility. Parasitology 132, 49–56 (2006).
Lu, W.-N., Chiu, M.-C. & Kuo, M.-H. Host life stage- and temperature-dependent density of the symbiont Buchnera aphidicola in a subtropical pea aphid (Acyrthosiphon pisum) population. J. Asia-Pac. Entomol. 17, 537–541 (2014).
Perrot-Minnot, M.-J., Guo, L. R. & Werren, J. H. Single and double infections with Wolbachia in the parasitic wasp Nasonia vitripennis effects on compatibility. Genetics 143, 961–972 (1996).
Van Opijnen, T. & Breeuwer, J. A. J. High temperatures eliminate Wolbachia, a cytoplasmic incompatibility inducing endosymbiont, from the two-spotted spider mite. Experim. Appl. Acarol. 23, 871–881 (1999).
Bordenstein, S. R. & Bordenstein, S. R. Temperature affects the tripartite interactions between bacteriophage WO, Wolbachia, and cytoplasmic incompatibility. PLoS ONE 6(12), e29106 (2011).
Jiang, R.-X. et al. The Influence of temperature and host gender on bacterial communities in the Asian citrus psyllid. Insects 12(12), 1054 (2021).
Hurst, G. D. D., Jiggins, F. M. & Robinson, S. J. What causes ineffcient transmission of male-killing Wolbachia in Drosophila?. Heredity 87, 220–226 (2001).
Hague, M. T. J. et al. Temperature effects on cellular host-microbe interactions explain continent-wide endosymbiont prevalence. Curr. Biol. 32(4), 878–88 (2021).
Sumi, T., Miura, K. & Miyatake, T. No seasonal trend in infection of the pale grass blue butterfly, Zizeeria maha (Lepidoptera: Lycaenidae), by Wolbachia. Appl. Entomol. Zool. 48, 35–38 (2013).
Sumi, T., Miura, K. & Miyatake, T. Wolbachia density changes seasonally amongst populations of the pale grass blue butterfly, Zizeeria maha (Lepidoptera: Lycaenidae). PLoS ONE 12(4), e0175373 (2017).
Cohen, C., Toh, E., Munro, D., Dong, Q. & Hawlena, H. Similarities and seasonal variations in bacterial communities from the blood of rodents and from their flea vectors. ISME J 9, 1662–1676 (2015).
Kriesner, P., Conner, W. R., Weeks, A. R., Turelli, M. & Hoffmann, A. A. Persistence of a Wolbachia infection frequency cline in Drosophila melanogaster and the possible role of reproductive dormancy. Evolution 70, 979–997 (2016).
Overholt, W. A., Diaz, R., Rosskopf, E., Green, S. J. & Overholt, W. A. Deep characterization of the microbiomes of Calophya spp. (Hemiptera: Calophyidae) gall-inducing psyllids reveals the absence of plant pathogenic bacteria and three dominant endosymbionts. PLoS ONE 10(7), e0132248 (2015).
Morrow, J. L., Hall, A. A. G. & Riegler, M. Symbionts in waiting: The dynamics of incipient endosymbiont complementation and replacement in minimal bacterial communities of psyllids. Microbiome 5(1), 1–23 (2017).
Morrow, J. L. et al. Characterization of the bacterial communities of psyllids associated with Rutaceae in Bhutan by high throughput sequencing. BMC Microbiol. 20(1), 1–6 (2020).
Shapoval, N. A., Nokkala, S., Nokkala, C., Kuftina, G. N. & Kuznetsova, V. G. The incidence of Wolbachia bacterial endosymbiont in bisexual and parthenogenetic populations of the psyllid genus Cacopsylla (Hemiptera, Psylloidea). Insects 12, 853 (2021).
Nakabachi, A., Inoue, H. & Hirose, Y. Microbiome analyses of 12 psyllid species of the family Psyllidae identified various bacteria including Fukatsuia and Serratia symbiotica, known as secondary symbionts of aphids. BMC Microbiol. 22(1), 1–21 (2022).
Kruse, A. et al. Combining ’omics and microscopy to visualize interactions between the Asian citrus psyllid vector and the Huanglongbing pathogen Candidatus Liberibacter asiaticus in the insect gut. PLoS ONE 12(6), e0179531 (2017).
Song, X. et al. Composition and change in the microbiome of Diaphorina citri infected with Candidatus Liberibacter asiaticus in China. Int. J. Trop. Insect. Sci. 39, 283–290 (2019).
Nachappa, P., Levy, J., Pierson, E. & Tamborindeguy, C. Diversity of endosymbionts in the potato psyllid, Bactericera cockerelli (Hemiptera: Triozidae), vector of zebra chip disease of potato. Curr. Microbiol. 62, 1510–1520 (2011).
Hail, D., Dowd, S. E. & Bextine, B. Identification and location of symbionts associated with potato psyllid (Bactericera cockerelli) lifestages. Environ. Entomol. 41, 98–107 (2012).
Cooper, W. R. et al. Wolbachia infection differs among divergent mitochondrial haplotypes of Bactericera cockerelli (Hemiptera: Triozidae). Ann. Entomol. Soc. Amer. 108, 137–145 (2015).
Fu, Z. et al. Host plants and Wolbachia shape the population genetics of sympatric herbivore populations. Evol. Appl. 13, 2740–2753 (2020).
Štarhová Serbina, L. et al. Microbiome of pear psyllids: a tale about closely-related species sharing their endosymbionts. Environ. Microbiol. https://doi.org/10.1111/1462-2920.16180 (2022).
Jarausch, B., Tedeschi, R., Sauvion, N., Gross, J. & Jarausch, W. Psyllid Vectors. In: Phytoplasmas: Plant Pathogenic Bacteria - II (eds. Bertaccini, A., Weintraub, P. G., Rao, G. P. & Mori, N.) 53–78 (Springer Singapore, 2019).
Lauterer, P. Results of the investigations on Hemiptera in Moravia, made by the Moravian museum (Psylloidea 2). Acta Musei. Morav. Sci. biol. 84, 71–151 (1999).
Hodkinson, I. D. Life cycle variation and adaptation in jumping plant lice (Insecta: Hemiptera: Psylloidea): A global synthesis. J. Natur. Hist. 43, 65–179 (2009).
Hague, M. T. J., Mavengere, H., Matute, D. R. & Cooper, B. S. Environmental and genetic contributions to imperfect wMel-like Wolbachia transmission and frequency variation. Genetics 215, 1117–1132 (2020).
Brinker, P., Fontaine, M. C., Beukeboom, L. W. & Falcao Salles, J. Host, symbionts, and the microbiome: the missing tripartite interaction. Trends Microbiol. 27, 480–488 (2019).
Sinkins, S. P. et al. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature 436, 257–260 (2005).
Ant, T. H., Herd, C., Louis, F., Failloux, A. B. & Sinkins, S. P. Wolbachia transinfections in Culex quinquefasciatus generate cytoplasmic incompatibility. Insect Mol. Biol. 29, 1–8 (2020).
James, A. C. & Ballard, J. W. O. Expression of cytoplasmic incompatibility in Drosophila simulans and its impact on infection frequencies and distribution of Wolbachia pipientis. Evol. 54, 1661–1672 (2000).
Koehncke, A., Telschow, A., Werren, J. H. & Hammerstein, P. Life and death of an influential passenger: Wolbachia and the evolution of CI-modifiers by their hosts. PLoS ONE 4(2), e4425 (2009).
Schuler, H. et al. Diversity and distribution of Wolbachia in relation to geography, host plant affiliation and life cycle of a heterogonic gall wasp. BMC Evol. Biol. 18(1), 1–5 (2018).
Bakovic, V., Schebeck, M., Telschow, A., Stauffer, C. & Schuler, H. Spatial spread of Wolbachia in Rhagoletis cerasi populations. Biol. Lett. 14(5), 20180161 (2018).
Kriesner, P., Hoffmann, A. A., Lee, S. F., Turelli, M. & Weeks, A. R. Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 9(9), e1003607 (2013).
Wolfe, T. M. et al. Comparative genome sequencing reveals insights into the dynamics of Wolbachia in native and invasive cherry fruit flies. Mol. Ecol. 30, 6259–6272 (2021).
Jerinić-Prodanović, D., Mihajlović, L. & Stojanović, A. Parasitoids of jumping plant-lice (Psylloidea, Hemiptera) from the family Encyrtidae (Hymenoptera, Chalcidoidea) in Serbia. Zootaxa 4577, 29–50 (2019).
Meyer, J. M. & Hoy, M. A. Molecular survey of endosymbionts in Florida populations of Diaphorina citri (Hemiptera: Psyllidae) and its parasitoids Tamarixia radiata (Hymenoptera: Eulophidae) and Diaphorencyrtus aligarhensis (Hymenoptera: Encyrtidae). Flor. Entomol. 91, 294–304 (2008).
Ashraf, H. J. et al. Comparative microbiome analysis of Diaphorina citri and its associated parasitoids Tamarixia radiata and Diaphorencyrtus aligarhensis reveals Wolbachia as a dominant endosymbiont. Environ. Microbiol. 24(3), 1638–1652 (2022).
Le Goff, G. J. et al. Effect of the instar of the pear psyllid Cacopsylla pyri (Hemiptera: Psyllidae) on the behaviour and fitness of the parasitoid Trechnites insidiosus (Hymenoptera: Encyrtidae). Eur. J. Entomol. 118, 279–287 (2021).
Tougeron, K. et al. Ecology and biology of the parasitoid Trechnites insidiosus and its potential for biological control of pear psyllids. Pest Manag. Sci. 77(11), 4836–47 (2021).
Dedeine, F., Ahrens, M., Calcaterra, L. & Shoemaker, D. D. Social parasitism in fire ants (Solenopsis spp.): A potential mechanism for interspecies transfer of Wolbachia. Mol. Ecol. 14, 1543–1548 (2005).
Gehrer, L. & Vorburger, C. Parasitoids as vectors of facultative bacterial endosymbionts in aphids. Biol. Lett. 8, 613–615 (2012).
Mascarenhas, R. O., Prezotto, L. F., Perondini, A. L. P., Marino, C. L. & Selivon, D. Wolbachia in guilds of Anastrepha fruit flies (Tephritidae) and parasitoid wasps (Braconidae). Genet. Mol. Biol. 39, 600–610 (2016).
Ossiannilsson, F. The Psylloidea (Homoptera) of Fennoscandia and Denmark. (Brill, E.J., 1992).
Burckhardt, D. Psyllid-key of Cacopsylla on Rosaceae. https://www.dlr.rlp.de/Psylliden-english (2010).
Braig, H. R., Zhou, W., Dobson, S. L. & O’Neill, S. L. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J. Bacteriol. 180, 2373–2378 (1998).
Baldo, L. et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72, 7098–7110 (2006).
Zhou, W., Rousset, F. & O’Neill, S. Phylogeny and PCR–based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. Lond. B 265, 509–515 (1998).
Jeffries, C. L. et al. Diverse novel resident Wolbachia strains in Culicine mosquitoes from Madagascar. Sci. Rep. 8(1), 1–5 (2018).
Zimmermann, B. L., Bouchon, D., Almerão, M. P. & Araujo, P. B. Wolbachia in Neotropical terrestrial isopods. FEMS Microbiol. Ecol. 91, (2015).
Kawasaki, Y., Schuler, H., Stauffer, C., Lakatos, F. & Kajimura, H. Wolbachia endosymbionts in haplodiploid and diploid scolytine beetles (Coleoptera: Curculionidae: Scolytinae): Wolbachia infection in scolytine beetles. Environ. Microbiol. Rep. 8, 680–688 (2016).
Schuler, H. et al. Wolbachia in parasitoids attacking native European and introduced Eastern cherry fruit flies in Europe. Environ. Entomoll 45, 1424–1431 (2016).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molec. Biol. and Evol. 35, 1547–1549 (2018).
Jolley, K. A., Bray, J. E. & Maiden, M. C. J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3, 124 (2018).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Molec. Biol. 215, 403–410 (1990).
Katoh, K., Rozewicki, J. & Yamada, K. D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 (2019).
Trifinopoulos, J., Nguyen, L.-T., von Haeseler, A. & Minh, B. Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucl. Acids Res. 44, W232–W235 (2016).
Ronquist, F. et al. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syste. Biol. 61, 539–542 (2012).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772–772 (2012).
Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE) 1–8 (IEEE, 2010).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucl. Acids Res. 49, 293–296 (2021).
Le Clec’h, W. et al. High virulence of Wolbachia after host switching: when autophagy hurts. PLoS Pathog. 8(8), e1002844 (2012).
Fox, J. & Weisberg, S. An R companion to applied regression 3rd edn. (Sage, 2019).
de Mendiburu, F. & Yaseen, M. Agricolae: statistical procedures for agricultural research. R package version 1.4.0. (2020).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing (2021).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016).
Acknowledgements
We thank two anonymous reviewers for providing the useful comments. LŠS is very grateful to her husband, Michal Štarha, for his enormous help in collecting psyllids in pear orchards. We also thank Dana Štarhová for providing access to pear trees in a family garden (locality CZ3), Stanislav Pekár for his continuous support during the field work and Ondřej Michálek for taking photographs of C. pyri. This work was supported by the Autonomous Province of Bozen-Bolzano to LŠS and HS, and by a Brno PhD Talent Scholarship, funded by the Brno City Municipality, to DG.
Author information
Authors and Affiliations
Contributions
L.Š.S, J.D. and H.S. conceived the project; L.Š.S. and D.G. collected the psyllid specimens; L.Š.S., I.M. and E.C. organised the data; L.Š.S. and J.D. conducted molecular analyses; J.D. and L.Š.S. performed statistical analyses; L.Š.S. prepared the first draft and wrote the manuscript together with J.D. and H.S. All authors provided feedback that helped to improve the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Štarhová Serbina, L., Gajski, D., Malenovský, I. et al. Wolbachia infection dynamics in a natural population of the pear psyllid Cacopsylla pyri (Hemiptera: Psylloidea) across its seasonal generations. Sci Rep 12, 16502 (2022). https://doi.org/10.1038/s41598-022-20968-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-022-20968-0
This article is cited by
-
Seasonal wild dance of dual endosymbionts in the pear psyllid Cacopsylla pyricola (Hemiptera: Psylloidea)
Scientific Reports (2023)
-
Variation in bacterial endosymbionts associated with Iranian populations of the common pistachio psylla, Agonoscena pistaciae (Hem.: Aphalaridae)
International Journal of Tropical Insect Science (2023)